December 25, 2025
The RBVI wishes you a safe and happy holiday season!
See our
2025 card and the
gallery of previous cards back to 1985.
December 16, 2025
The ChimeraX 1.11 production release is
available! See the
change log
for what's new.
November 21, 2025
The ChimeraX 1.11 release candidate is
available –
please try it and report
any issues. See the
change log
for what's new.
This will be the last release to support Red Hat Enterprise Linux 8 and
its derivatives.
Previous news...
UCSF ChimeraX
UCSF ChimeraX (or simply ChimeraX)
is the next-generation molecular visualization program from the
Resource for Biocomputing,
Visualization, and Informatics (RBVI),
following UCSF Chimera.
ChimeraX can be downloaded free of charge
for academic, government, nonprofit, and personal use.
Commercial users, please see
ChimeraX commercial licensing.
ChimeraX is developed with support from National Institutes of Health R01-GM129325.
 ChimeraX on Bluesky:
@chimerax.ucsf.edu
ChimeraX on Bluesky:
@chimerax.ucsf.edu
A cryoelectron microscopy map of the 26S proteasome
(EMD-4321) is shown at the author-recommended contour level
in two different lighting modes: “simple” on the left
and “soft” on the right.
Soft lighting includes ambient lighting and shadowing (occlusion)
and can be turned on with the command
lighting soft
or by clicking the
Graphics
icon
 .
.
For setup of the righthand image,
see the command file ambient.cxc.
More features...
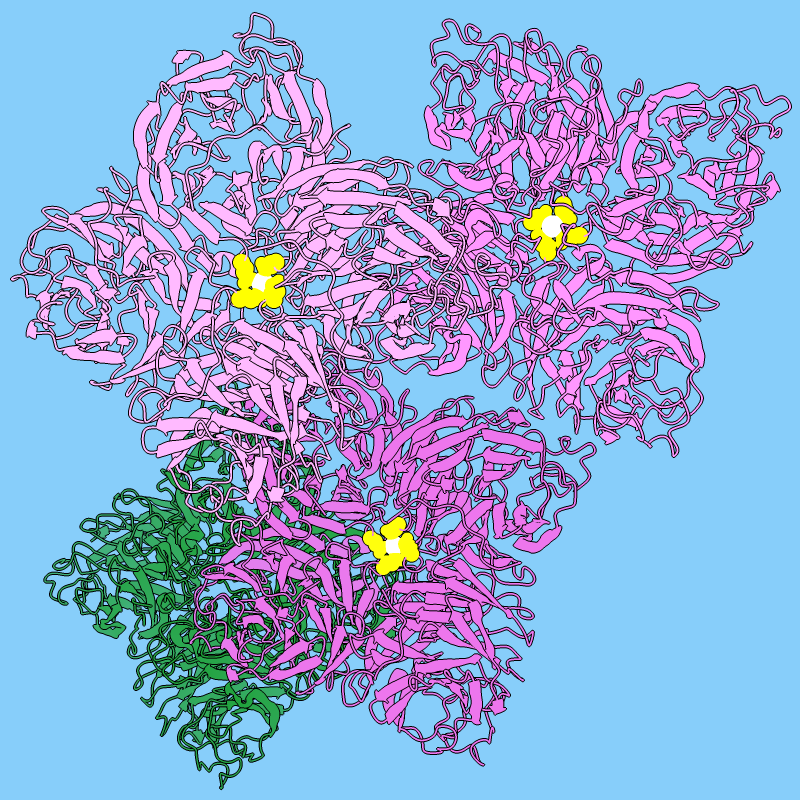
Influenza neuraminidase is an enzyme that promotes the spread of influenza
virus among host cells.
It is the target of oseltamivir and related antiviral drugs.
The image shows tetramers of neuraminidase (PDB
3k3a) styled as flowers. Three tetramers are in different
shades of pink, with a central metal ion in white and nearby residues in yellow,
and a fourth tetramer is colored green to resemble leaves.
Each monomer or “petal” is a six-bladed β-propeller.
For image setup other than orientation,
see the command file flowers.cxc.
The Chimera Image Gallery includes a similar image.
More images...