

 about
projects
people
publications
resources
resources
visit us
visit us
search
search
about
projects
people
publications
resources
resources
visit us
visit us
search
search
Quick Links
Featured Citations
An ATP-gated molecular switch orchestrates human mRNA export. Hohmann U, Graf M et al. Nature. 2026 Jan 22;649(8098):1042–1050.
ZAK activation at the collided ribosome. Huso VL, Niu S et al. Nature. 2026 Jan 22;649(8098):1051–1060.
Semantic design of functional de novo genes from a genomic language model. Merchant AT, King SH et al. Nature. 2026 Jan 15;649(8097):749-758.
Structural basis of regulated N-glycosylation at the secretory translocon. Yamsek M, Ma M et al. Nature. 2026 Jan 15;649(8097):777–784.
Top-down scoring of spectral fitness by image analysis for protein structure validation. Harding BD, DeZonia B et al. J Chem Inf Model. 2026 Jan 12;66(1):567-576.
More citations...News
December 25, 2025

|
December 16, 2025
The ChimeraX 1.11 production release is available! See the change log for what's new.
November 21, 2025
The ChimeraX 1.11 release candidate is available – please try it and report any issues. See the change log for what's new. This will be the last release to support Red Hat Enterprise Linux 8 and its derivatives.
Previous news...Upcoming Events
UCSF ChimeraX (or simply ChimeraX) is the next-generation molecular visualization program from the Resource for Biocomputing, Visualization, and Informatics (RBVI), following UCSF Chimera. ChimeraX can be downloaded free of charge for academic, government, nonprofit, and personal use. Commercial users, please see ChimeraX commercial licensing.
ChimeraX is developed with support from National Institutes of Health R01-GM129325.
 ChimeraX on Bluesky:
@chimerax.ucsf.edu
ChimeraX on Bluesky:
@chimerax.ucsf.edu
Feature Highlight
 |
 |
 |
 |
 |
 |
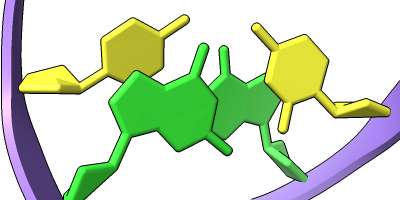
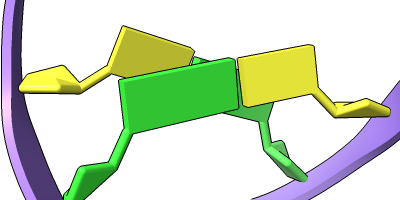
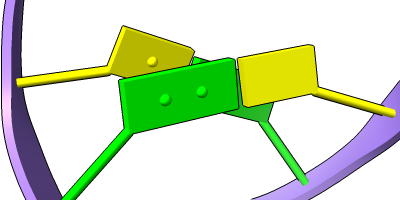
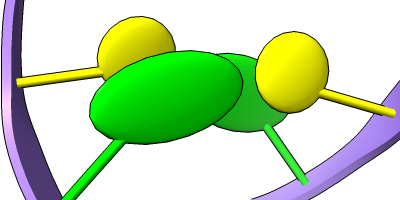
Different representations of nucleotides can be shown with the nucleotides command or Toolbar icons. Options include filled rings, slabs for bases (box, muffler, or ellipsoid shape), bumps on slabs to show base orientation, simple tubes instead of ribose atoms, and continuous or broken ladder rungs. Nucleotide representations can be the same color as the ribbon or a different color, and multiple nucleotide styles can be used within a single structure.
See also: Presets menu
More features...
Example Image
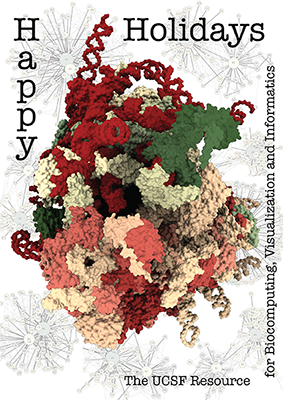
The architecture of the human ribosome has been determined at near-atomic resolution by electron microscopy (Anger et al., Nature 497:80 (2013)). The structure, comprising 82 proteins and five RNA molecules, is shown with shadows cast from all directions to accentuate depth. In the background are schematic representations of contacts between the component molecules.
See the image setup script card.cxc using the 'Tis the Season color palette (credit to MrsP). See also the RBVI holiday card gallery.
About RBVI | Projects | People | Publications | Resources | Visit Us
Copyright 2018 Regents of the University of California. All rights reserved.