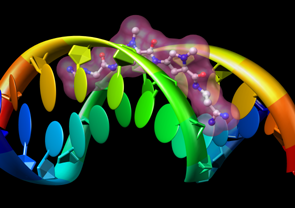
|
Molecular Graphics
- interactively manipulable stick, ball-and-stick, CPK, ribbon,
and special nucleotide
representations;
molecular surfaces
- highly intuitive translation, scaling, and rotation;
Side View
tool for adjusting clipping planes and scaling
- interactive color editing
in various color spaces (RGB, CMYK, etc.), including
transparency
- ability to save high-resolution images for presentation
and publication
- stereo viewing (side-by-side and time-sequential)
|
|
|
Chemical Knowledge
|
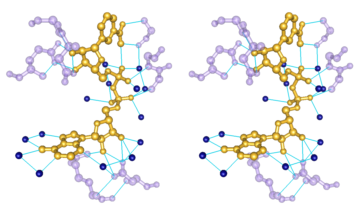
|
| |
- determination of
atom types in arbitrary molecules, including non-standard residues
- ability to add hydrogen atoms
- high-quality hydrogen bond identification
- selection of atoms/bonds by element, atom type,
functional group, amino acid category
- interactive bond rotation, distance and angle measurements
|
|
|

|
Sequence Viewer
The Multalign Viewer
extension displays multiple sequence alignments, calculates
and shows a consensus sequence and conservation
histogram, and allows regions to be defined and colored.
 Structures opened in Chimera are automatically associated with a
matching sequence. Structures can be superimposed based on
the sequence alignment, and actual/predicted secondary
structure features can be depicted on the alignment. When a
region is defined by dragging within the sequence, the corresponding
residues of the structure are highlighted. The color swatches
behind the sequence names match the corresponding structures.
Structures opened in Chimera are automatically associated with a
matching sequence. Structures can be superimposed based on
the sequence alignment, and actual/predicted secondary
structure features can be depicted on the alignment. When a
region is defined by dragging within the sequence, the corresponding
residues of the structure are highlighted. The color swatches
behind the sequence names match the corresponding structures.
|
|
|
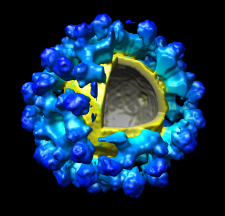
Volume Data
Chimera can display
three-dimensional electron and light microscope data,
x-ray density maps, electrostatic potential and other volumetric
data. Contour surfaces, meshes and volumetric
display styles are provided and thresholds can be changed
interactively. Maps can be
colored, sliced, segmented,
and modifications can be saved.
Markers can be placed and structures can be traced.
The accompanying image shows a density map of
Kelp fly virus from electron microscopy colored radially and
with an octant cut out.
|
|
|
|
|
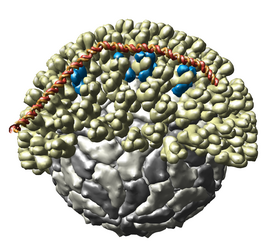
Multiscale Models
The Multiscale
extension allows Chimera to display large complexes such as
virus capsids,
ribosomes, and
chromatin.
It displays quaternary structure of PDB models and allows you
to select subunits and show atomic resolution detail. Matrices
are read from PDB files that specify the biological unit.
Crystallographic packing can also be shown.
|
|
|
Molecular Dynamics / Lenses
Chimera can
display
molecular dynamics trajectories in a variety of formats:
AMBER,
CHARMM,
GROMACS,
GROMOS,
MMTK,
NAMD,
PDB, and
X-PLOR.
All normal Chimera analysis and display capabilities
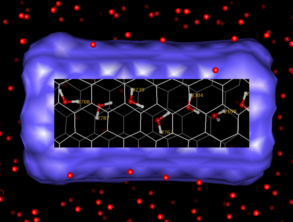
are available with trajectories. One such capability is to place
"lenses" on the screen: rectangular regions containing
markedly different display properties. The accompanying
image shows one trajectory frame of a buckytube immersed in water.
A lens has been placed over the center of the buckytube to reveal
the hydrogen-bonded chain of waters in the center of the tube, along
with their residue numbers. The simulation was done with polarizable
molecular dynamics using the Amber8 Sander module, and provided
courtesy of J. Caldwell.
|
|
|
|
|
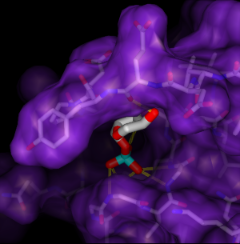
Screening Docked Molecules
The program DOCK
calculates possible binding orientations,
given the structures of "ligand" and "receptor" molecules.
Typically, a large database of small molecule structures is searched
for compounds that may bind the receptor. The Chimera extension
ViewDock
facilitates interactive selection of promising compounds
from the output of DOCK. The molecules can be viewed in the context
of the binding site and optionally, screened by number of hydrogen bonds
to the receptor.
The Dock Prep extension
prepares a receptor for input to a docking program by
adding hydrogens, assigning partial charges, and performing other
related tasks.
|
|
|
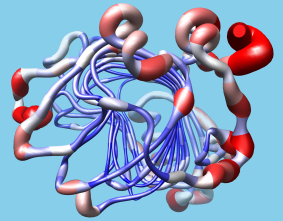
User-Driven Analysis
Users can easily import structure-related data into Chimera
in the form of "attributes,"
i.e. values associated with atoms, residues, or models.
The data can be imported with the
Define Attribute
extension and then visually
represented as color ranges, varying atom sizes,
or "worm" backbone depictions, using the
Render by Attribute
extension.
Such data can also be
manipulated programmatically in Chimera, and in fact Chimera
was designed with extensibility and programmability in mind.
It is largely implemented in Python, with certain features
coded in C++ for efficiency.
Python is an easy-to-learn
interpreted language with object-oriented features. All of Chimera's
functionality is accessible through Python and users can implement
their own algorithms and extensions without any Chimera code changes,
so any such custom extensions will continue to work across
Chimera releases. Many
programming examples
are provided to assist extension writers.
|
|
|
|
More Details
For more details on these and other Chimera capabilities,
please see our
examples page.
|


 home
overview
research
resources
outreach & training
outreach & training
visitors center
visitors center
search
search
home
overview
research
resources
outreach & training
outreach & training
visitors center
visitors center
search
search


 home
overview
research
resources
outreach & training
outreach & training
visitors center
visitors center
search
search
home
overview
research
resources
outreach & training
outreach & training
visitors center
visitors center
search
search
