Preferences
Choosing Settings... Preferences from the menu opens the
Preferences Tool.
Options within are grouped by Category:
By default, preferences are stored in the file
$HOME/.chimera/preferences, where $HOME refers to
the user's home directory. The location can be changed in the
Preferences section
of the Preferences Tool.
The preferences file can contain information in addition to
the Preferences Tool settings,
including startup and icon preferences from the
Extension Manager
and names of recently accessed files and directories.
Save saves the current Preferences Tool settings to the
preferences file.
Restore replaces the current settings with
those already specified in the file, whereas Reset replaces the
current settings with the defaults.
Help opens this manual page in a browser window.
Close dismisses the Preferences Tool.
Default settings are indicated below in bold.
Background
The Background section of the
Preferences Tool includes:
- Background color (a
color well) - the color is reported
as "No" if the background color has not been changed, indicating
that the default (black) is being used
- Selection highlight method (outline/fill) - how
a selection is highlighted in the graphics window
- Selection highlight color (a
color well, green by default) -
what color to use for highlighting a selection
in the graphics window
- Draw lens borders (true/false) - whether to draw outlines
of any lenses in the graphics window
- Lens border color (a
color well, green by default)
- what color to use for outlining any
lenses in the graphics window
General
The General section of the
Preferences Tool includes:
- Confirm exit (true/false) - whether to ask for
confirmation after Session... Exit has been chosen from the menu
- Show balloon help (true/false) - whether to show
balloon help,
explanatory text that pops up when the cursor is left in certain positions
- Show atomspec balloon
(true/false) - whether to show
label information when the cursor is left on top of an atom or bond.
- Atomspec display style
(simple/Midas atomspec/Midas-dependent) -
whether to describe atoms with a simple style that consists of residue
name, residue number, and atom name, or with a Midas
atomspec string;
the default is to use the simple style when the
Midas Emulator is not shown
and atomspec strings
when the Midas Emulator
is shown (affects atomspec balloons and the
Structure Measurements panels)
- File lists path style (file - leading path/full path) -
how to list files and their locations in certain dialogs (such as
Files to read at startup within the Midas
section of the Preferences Tool); a change
will not be evident until after Chimera is restarted
- Confirm starting extension (true/false) - whether to ask
for confirmation before starting certain extensions (for example,
MainChain
or AddH)
- Toolbar visible (no/yes/auto) - whether or not to display
the tool bar,
where icons for extensions are placed; the auto option
only shows the tool bar if it contains one or more icons
- Toolbar side (left/right/top/bottom) - position of the
tool bar relative to the graphics window
- Use short open/save browser (true/false) - whether
open/save dialogs
should show only the two lowest directory levels
(more details)
- Open dialog starts in directory from last session
(true/false) - whether the first
open/save dialog invoked during a session should start in the directory
where a file was last opened or saved in the previous session, or in the
current working directory. The default is true on
Windows systems, false on the others.
Image Credits
The Image Credits section of the
Preferences Tool specifies information to be included
in saved TIFF image files. This information does not affect the image itself.
- Artist (user's login name by default) - optional, name of the
image creator
- Copyright (blank by default) - optional, a copyright statement
Messages
The Messages section of the
Preferences Tool controls the disposition of
command, status, warning, and error messages.
- Show replies at startup (true/false) - whether to
open the Reply Log upon Chimera startup
- Show status line (true/false) - whether to
show a line for status messages under the main
Chimera window (otherwise, status messages are not visible);
can also be set from the
Accelerators dialog
- Clear status line after (5 seconds/10 seconds/20 seconds/30
seconds/1 minute/never) - how long a status message will be shown in
the status line (when not overwritten by subsequent status messages);
applies to status messages appearing in certain dialogs as well as
the main Chimera status line
- Command (reply log only/dialog) - whether command messages
should appear in the reply log only, or also generate a dialog box
- Warning (reply log only/dialog) - whether warnings
should appear in the reply log only, or also generate a dialog box
- Error (reply log only/dialog) - whether error messages
should appear in the reply log only, or also generate a dialog box
Midas
The Midas section of the
Preferences Tool is only available when the
Midas Emulator
is shown.
Files to read at startup refers to what files, if any,
contain commands to be executed when (or if) the
Midas Emulator is shown.
There are two defaults:
- $HOME/.chimera/midasrc (in a directory named .chimera in
the user's home directory, here referred to as $HOME)
- .chmidasrc in the current directory
and either or both are executed (in the order listed), if found.
The defaults can be deleted and/or additional names and locations
can be added to the list.
The files are treated as if they had been
sourced in the
Midas Emulator.
Whether files are listed by name followed by path or with
the path preceding the name is controlled in the
General preferences.
The midasrc mechanism is similar to that in MidasPlus, where
a file named .midasrc in the user's home directory was automatically
executed upon program startup. In general, the same file can be used
here after commands unsupported by Chimera
have been removed.
A midasrc-type file can be created by manual text-editing or by
using Record... in the Command History
tool to save commands already used in Chimera.
Mouse
By default, structures are rotated with the left mouse button
and XY-translated with the middle mouse button (for more
details, see mouse manipulation).
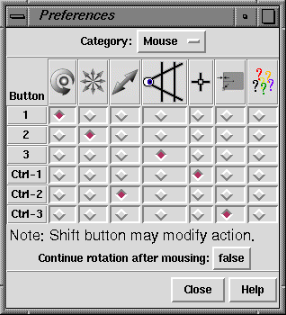 Mouse button functions can be reassigned within the Mouse section
of the Preferences Tool.
This panel may be used to assign functions to the left, middle,
and right mouse buttons (Buttons 1, 2, and 3, respectively).
Ctrl-1, Ctrl-2, and Ctrl-3 indicate the functions of the left, middle, and
right mouse buttons when the Ctrl key is also held down.
Current assignments are indicated by color-highlighted checkboxes.
Since a button can only have one function,
selecting a function by mouseclick turns off the previously selected
function in that row. The figure shows the default assignments.
Going from left to right,
the symbols across the top of the panel signify rotation,
XY-translation, Z-translation, scaling, atom and bond
picking
(selection),
lens control,
and a pop-up menu bar when the standard menu bar is not
visible (not implemented).
Mouse button functions can be reassigned within the Mouse section
of the Preferences Tool.
This panel may be used to assign functions to the left, middle,
and right mouse buttons (Buttons 1, 2, and 3, respectively).
Ctrl-1, Ctrl-2, and Ctrl-3 indicate the functions of the left, middle, and
right mouse buttons when the Ctrl key is also held down.
Current assignments are indicated by color-highlighted checkboxes.
Since a button can only have one function,
selecting a function by mouseclick turns off the previously selected
function in that row. The figure shows the default assignments.
Going from left to right,
the symbols across the top of the panel signify rotation,
XY-translation, Z-translation, scaling, atom and bond
picking
(selection),
lens control,
and a pop-up menu bar when the standard menu bar is not
visible (not implemented).
Continue rotation after mousing (true/false) indicates whether
to rotate continuously when the mouse button assigned to rotation is released
while the cursor is in motion.
New Molecules
The New Molecules section of the
Preferences Tool controls how newly opened molecules
will initially appear:
- use new color for each model (true/false) -
set the model-level color
(see coloring hierarchy)
differently for each new molecule model;
submodels with
the same main model number will be assigned the same model-level color.
This preference can also be controlled with the commands
set/~set autocolor.
- otherwise, use color (a
color well, by default showing
a light gray color) -
color at the model level, see coloring hierarchy
- ribbon depiction (none/flat ribbon/edged
ribbon/rounded ribbon) - type of secondary structure
ribbon shown (see also resrepr).
Ribbons are only drawn for proteins and nucleic acids.
Protein secondary structure is determined from HELIX and SHEET records
in the input file, or if these are not present, using
ksdssp.
- line width (1 by default)
- pixel width of lines drawn to show bonds in the wire draw mode
- atom style (dot/ball/endcap/sphere) - atom
draw mode
- bond style (wire/stick) - bond
draw mode
- stick diameter (0.2 by default)
- angstrom thickness of bonds in the stick draw mode
- ball size (0.25 by default)
- fraction of VDW radius
used for ball radius when atoms are in the ball draw mode
- coloring by element (true/false) - whether
byatom coloring is used;
this overrules the model-level color above, see
coloring hierarchy
- auto-chaining (off/on)
- whether to connect atoms that precede and follow undisplayed segments
(whether to draw pseudobonds between them)
PDB
The PDB section of the
Preferences Tool includes:
- Fetch from web as necessary (true/false) -
whether to retrieve an input PDB file from the
Protein DataBank web site
when it is not found locally
- Personal PDB directories - additional directories to look
in for PDB files; the user may Add a directory to the list of places to look,
or choose a previously added directory from the list and then Delete it from
the list
Page Setup
The Page Setup section of the
Preferences Tool sets specifications for images
saved to TIFF files using the Image Panel.
This section can also be invoked from the
Image Panel.
- Units
(centimeters/inches/millimeters/points)
- the units used for describing the options below
- Printer Resolution - values in this section should be compatible
with the specifications of the printer used
- Horiz. dots per unit (by default, 400 per inch
or the equivalent in other units)
- Vert. dots per unit (by default, 400 per inch
or the equivalent in other units)
- Colors per dot (3/3+black/4/5/5+black/6)
- Shades per color (2 by default)
Preferences
The Preferences section of the
Preferences Tool specifies:
Location - where the preferences file for the current session
should be located:
- preferences (in the current directory)
- .chimera/preferences (in a directory named .chimera in the current
directory
- $HOME/.chimera/preferences (in a directory named .chimera in
the user's home directory, here referred to as $HOME)
 Mouse button functions can be reassigned within the Mouse section
of the Preferences Tool.
This panel may be used to assign functions to the left, middle,
and right mouse buttons (Buttons 1, 2, and 3, respectively).
Ctrl-1, Ctrl-2, and Ctrl-3 indicate the functions of the left, middle, and
right mouse buttons when the Ctrl key is also held down.
Current assignments are indicated by color-highlighted checkboxes.
Since a button can only have one function,
selecting a function by mouseclick turns off the previously selected
function in that row. The figure shows the default assignments.
Going from left to right,
the symbols across the top of the panel signify rotation,
XY-translation, Z-translation, scaling, atom and bond
picking
(selection),
lens control,
and a pop-up menu bar when the standard menu bar is not
visible (not implemented).
Mouse button functions can be reassigned within the Mouse section
of the Preferences Tool.
This panel may be used to assign functions to the left, middle,
and right mouse buttons (Buttons 1, 2, and 3, respectively).
Ctrl-1, Ctrl-2, and Ctrl-3 indicate the functions of the left, middle, and
right mouse buttons when the Ctrl key is also held down.
Current assignments are indicated by color-highlighted checkboxes.
Since a button can only have one function,
selecting a function by mouseclick turns off the previously selected
function in that row. The figure shows the default assignments.
Going from left to right,
the symbols across the top of the panel signify rotation,
XY-translation, Z-translation, scaling, atom and bond
picking
(selection),
lens control,
and a pop-up menu bar when the standard menu bar is not
visible (not implemented).