

The goal of this project is to determine how the 30 nm chromatin fiber is packaged in a condensed Drosophila chromosome.
The figures below summarize some of the visualizations described in more detail in:
The first 8 figures are attempts at revealing structure in the electron microscope chromosome data using 3D image processing ideas. None of the methods were very successful. A more powerful approach for extracting information from very noisy data is to fit a model. The last figures show such a model.

|
Figure 1:
A slice through a Drosophila chromosome shown with Priism program. The region in the red box is shown in the next figure. The three dimensional data was calculated from transmission electron microscope images collected by Maria de Sousa Vieira formerly a researcher in John Sedat's group. The specimen is tilted over a range of angles and a 3D intensity matrix is computed by back-projection. |

|
|
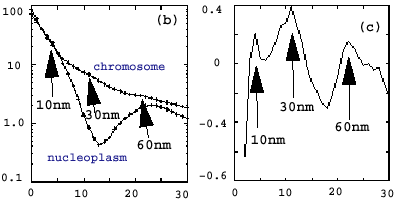
| Figure 2: Maria used spectral analysis techniques to show that there were 30 nm features in the chromosome that were not present in the non-chromosome regions of the data. Spectral bumps at other relevant length scales were detected. I have only a couple pages (1, 2) written by Maria on her results. | |

|

|
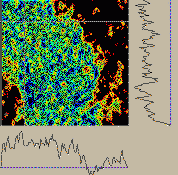
Figure 3:
Stereo pair isosurface of electron microscope tomography for boxed region from figure 1 shown with Chimera. Quicktime movie (3.3 Mb) of rotating volume representation of chromosome data. |
|
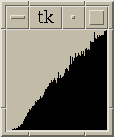
Figure 4:
Contour plot of 2D sections of electron microscope data using NMR spectrum display program Sparky. Plots of 1D sections of data corresponding to white crosshair are shown along edges. Quicktime movie flipping through the 64 z-axis planes. |
|
Figure 5:
Histogram of pairwise separations of peaks in data. Separations range from 0 to 15 nm in the figure. Semi-regular packing of nucleosomes in the 30 nm chromatin fiber might show up as peaks in this histogram. |
|
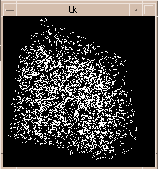
Figure 6:
Line segments connecting all pairs of peaks in electron microscope data set separated by 9 to 9.5 nm. If this is a prominent nucleosome spacing in the 30 nm chromatin fiber the fiber path might be visible. Not much can be seen rotating this hay stack of 3854 lines segments. |

|
Figure 7:
Ridges (yellow) and valleys (blue) in electron microscope data. A ridge is defined as a point where the data value is less at most neighboring points. The neighbors are the 26 data values in a 3 by 3 by 3 cube centered on the point. The connectivity patterns seen in the chromosome and nucleoplasm regions of the data are similar. So this is unlikely to reveal structure of the chromosome. |
|
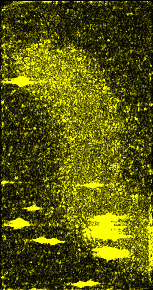
Figure 8:
Absolute deviation from a 15 nm spatial average. It is not known what features of the chromosome will be most prominent with the uranyl acetate staining that was used. John Sedat was interested in whether changes in the local standard deviation of data values might reveal structures. |
|
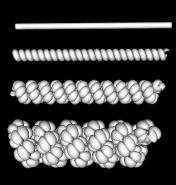
Figure 9:
Model of 30 nm chromatin fiber wrapped in 3 level helix. The fiber is shown as a tube at the top. It is wrapped into a simple helix in the second model from the top. That helix is wrapped into a helix in the third model from the top. The bottom most model is a 3rd level helix. The model is physically implausible because of the high compression near the center of the helices. |
|
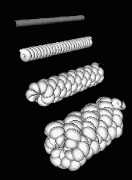
Figure 10:
The helix model of figure 9 is shown from an angle. The bottom most model shows rows of bumps in lines that make the structure appear to consist of multiple strands wound around each other. This surface appearance arises from the staggering of turns in the finer two levels of helices. Some light microscope data hints at the highest level packaging being double helical. |
Laboratory Overview | Research | Outreach & Training | Available Resources | Visitors Center | Search