bioCycPlugin: Cytoscape plugin for loading pathways from Pathway Tools web services

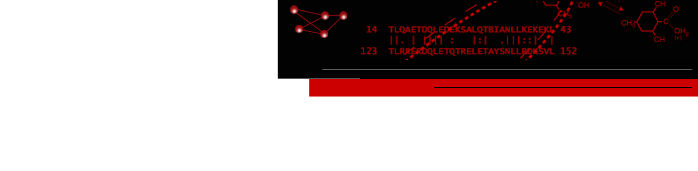
Figure 1. bioCycPlugin in action. In this example we've loaded the glycolysis I network from the HumanCyc database at http://websvc.biocyc.org/. We loaded the network and then used the chemViz plugin to show the 2D structures for AMP, ADP, and ATP.
The bioCycPlugin is a Cytoscape plugin that utilizes the web services infrastructre in Cytoscape to access web services using Pathway Tools version 14.5 or better. The plugin allows users to select the site they want to use (the BioCyc repostory is the default). The plugin provides the user with a list of databases to choose from. The user's repostory and database selections are saved in the user's home folder. The user may search the database by entering the name of a pathway, protein, or compound. They will then be presented with a list of pathways containing that term. Double-clicking on the desired pathway will load it into Cytoscape using the BioPAX format. As part of the load process, the SMILES strings for any small molecules with structures are pulled out of the CML and put into a "smiles" cytoscape attribute to provide compatability with chemViz. The bioCycPlugin depends on version 2.8 of Cytoscape and is available from the Cytoscape plugin manager.
Laboratory Overview | Research | Outreach & Training | Available Resources | Visitors Center | Search