SSD 
BACKGROUND
The SSD extension of
Chimera
provides a user interface to the
Structure Superposition Database (SSD).
The database contains structural superposition data
from a large number of pairwise structure comparisons using the program
MinRMS. Each pairwise comparison yields a series
of superpositions ranging from few to many pairs of alpha-carbons matched.
For each number of alpha-carbons paired, the superposition
with the lowest root-mean-square deviation (RMSD) is kept.
The SSD extension is not distributed with Chimera,
but it is freely available from the
Structure Superposition Database web site.
Using the SSD extension, users can view all or a subset of the
superpositions for a given pair of structures, view multiple structure
superpositions (when pairwise comparisons have been done for a given
structure versus multiple other structures), access RMSD values
and other quantities describing the superpositions, and view pairwise and
multiple sequence alignments derived from the structural superpositions.
Beginning users of Chimera should consult the tips.
STARTUP
SSD should be listed in the Chimera menu under
Tools... Structure Comparison. If SSD is not listed:
- Make sure that the module has been
downloaded
and unzipped; this will make a directory (folder) named
ssd Folder that contains the directories ssd and AlignPlot.
- Make sure that Chimera can find the module.
This can be done in either of two ways:
- Move or copy these two directories (ssd and AlignPlot)
into the share directory of your Chimera installation.
- Add the directory containing these two directories
to the list of places where Chimera should look:
- in Chimera, choose Favorites... Add to Favorites/Toolbar.
- Add the location. For example,
add /Users/meng/Desktop/ssd Folder
if it contains the ssd and AlignPlot directories.
- Save this setting so that SSD will automatically
appear in the menu in later uses of Chimera.
Choose Tools... Structure Comparison... SSD to show the SSD Browser,
the graphical interface to the database.
It includes a main panel and some smaller sections:
The Display Options
control which superpositions are displayed and
how they are colored.
The buttons Back and Forward can be
used to move among previous browser contents.
Mainchain shows the mainchain atoms only of any displayed structures.
Close closes the SSD Browser, and Help opens this
manual page in a web browser.
CHOOSING STRUCTURES
At least two structures must be chosen before superpositions can be viewed.
Even when several structures are chosen, there is another step
(choosing superpositions)
to indicate which of the following will be examined:
- all or a subset of the superpositions between two structures
- a single superposition of multiple structures
There are several ways to start choosing structures from the database.
The Browse superfamilies button Go
brings up a list of structures in the SSD.
It may take a few seconds for the entire list to appear.
Structures are listed by 4-character PDB identifier and arranged
by superfamily as defined in SCOP.
Clicking a PDB identifier displays the structure in the graphics window
and information on the structure
in the SSD Browser.
The Back button can be used to return to the list.
Activating a checkbox next to an individual PDB identifier chooses
that structure; activating a checkbox next to a superfamily name
chooses all structures in the superfamily.
Clear all deactivates all checkboxes.
After at least two structures have been chosen, pressing View
superpositions brings up the dialog for
choosing superpositions.
The Search structures section
allows searching by a single PDB identifier.
If the structure is not in the SSD, a message that no proteins
were found will appear. When the PDB identifier
(uppercase or lowercase) of a structure in the SSD
is entered and Search is pressed, the structure is displayed
in the graphics window and
information on the structure is shown in the SSD Browser.
The individual structure information includes protein name and source
species. To the right of the PDB identifier is a checkbox that toggles
model display and a menu for changing the model color.
Pressing Select superposition partner(s) brings up a
list of structures in the SSD. After at
least one other structure has been chosen, pressing View
superpositions brings up the dialog for
choosing superpositions.
The Search superpositions
section provides the most direct route to
viewing pairwise superpositions. Entering the PDB
identifiers of two structures in the SSD that have been superimposed
with each other and pressing Search bypasses the additional step
for choosing superpositions and
goes straight to the pairwise superposition view.
If the SSD does not contain superpositions between the two structures,
a message that no superpositions were found will appear.
The Search multiple structures section
allows a list of PDB identifiers to be entered. After Search
is pressed, PDB identifiers not
found in the database are reported; if none of the structures
are in the SSD, an additional error message will appear.
If some of the structures are in the SSD,
the dialog for choosing superpositions
will appear.
CHOOSING SUPERPOSITIONS
After structures have been chosen,
it is usually still necessary to choose superpositions;
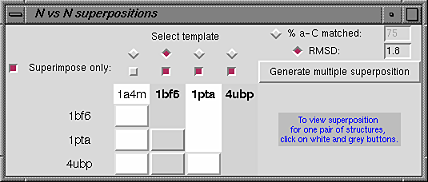
this is done within the N vs N superpositions dialog.
To choose all or a subset of the superpositions between
two structures (a pairwise
superposition), click on the white or gray
button in the row and column corresponding to the desired pair of
structures. These buttons are in the lower part of the
N vs N superpositions dialog.
None of the other settings
in the dialog affect what is obtained by clicking in this checkerboard
area. The Display Options section of
the SSD Browser controls whether all or a subset of the
pairwise superpositions will be shown. To view only a subset
based on user-specified ranges for RMSD and percent alpha-carbons matched,
specify the ranges before clicking the white or gray button.
To choose a single superposition of multiple structures
(a multiple superposition):
- specify which structure should be treated as the template;
the set of superpositions of each additional structure with the template
will be examined
- specify a criterion for picking a single superposition from each
set of superpositions:
- % a-C matched - for example, if 75% and
the shorter protein has 400 alpha-carbons, indicates
the superposition with 300 alpha-carbons matched
- OR -
- RMSD - for example, if 2.0 angstroms, indicates
the superposition with the set of alpha-carbons matched
at 2.0 angstroms RMSD
If all of the structures listed in the
N vs N superpositions dialog
are to be included, click Generate multiple superposition
to display the multiple superposition and
associated information.
If only a subset of the structures listed in the
N vs N superpositions dialog
are to be included, activate the Superimpose only checkbox,
deactivate the checkbox above each structure that should not be included,
and then click Generate multiple superposition
to display the multiple superposition and
associated information.
PAIRWISE SUPERPOSITIONS
When superpositions between two structures have been
chosen, the two structures
are displayed in the Chimera window and
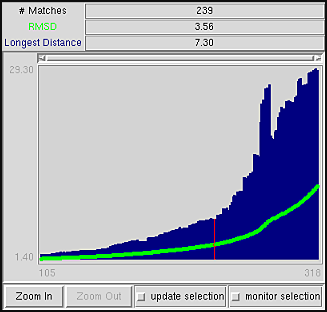
a graph
of the number of alpha-carbons matched versus RMSD is
shown in the SSD Browser.
Above the graph are the two structures'
PDB identifiers, checkboxes that toggle model display, and menus
for changing model color.
The graph depicts all (or a specified subset) of the superpositions calculated
for a pair of structures. The number of paired alpha-carbons, or matches,
is plotted versus RMSD (green) and the longest distance between paired
alpha-carbons (blue).
Clicking with the mouse within this graph determines
which superposition is displayed in the graphics window.
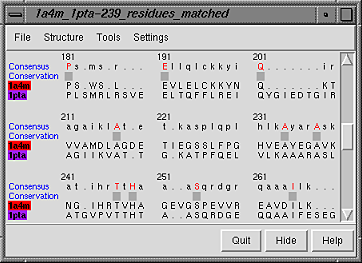
In the figure, the graph has been clicked to choose the superposition
with 239 matches, an RMSD value of 3.56 angstroms, and a
longest distance between matched alpha-carbons of 7.30 angstroms.
The superposition is marked by a vertical red line on the graph.
Zoom In expands the X axis so that the graph shows a smaller
range of match values in the same amount of space; Zoom Out
reverses this. When the graph is zoomed in, the entire range of match
values can still be viewed using the slider above the graph.
The
Display Options section of the SSD Browser only pertains
to pairwise superpositions. One can:
- view all superpositions - view the entire set of superpositions
calculated for the pair of structures
- OR -
- view superpositions in range - view the subset of superpositions
calculated for the pair of structures that fall within
the specified ranges of both:
- RMSD - RMSD of the sets of matched alpha-carbons
- % a-C matched - percent of alpha-carbons matched relative
to the number of residues in the shorter protein
Currently, to have an effect,
the ranges must be set before a pairwise superposition
is chosen.
The Color by options are:
- structure - make each structure a single color, as shown by
its color setting
- matched/unmatched - color the matched parts of a structure
according to its color setting,
and color the unmatched parts gray
Changing the coloring option does not change the display immediately,
but affects subsequently displayed superpositions.
Clicking the Transform Matrix button next to the
graph opens a small window showing the transformation matrix used for the
current superposition. The window title is of the form
pdb1 vs pdb2 - N residues matched
The matrix is that which transforms pdb2 onto
pdb1 in the current superposition, which
has N alpha-carbon pairings.
Clicking Sequence Align opens a
structure-based sequence alignment corresponding to the
current superposition, using the Chimera extension
Multalign Viewer.
In the structure-based sequence alignment, only the residues matched in
the superposition are aligned in the sequence; unmatched residues are treated
as insertions (gaps in the other sequence).
For details on the sequence display and how it can be customized, see the
Multalign Viewer documentation. Briefly,
the name of each sequence is shown in bold
over a rectangle matching the model color. This indicates
the sequence is associated with the structure shown in the
Chimera window.
MULTIPLE SUPERPOSITIONS
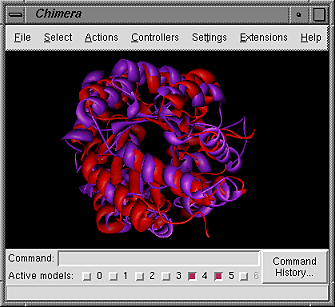
When a superposition of multiple structures has been
chosen, the superpositions
(plural since a multiple superposition is actually a combination of
pairwise superpositions specified by some criterion)
are displayed in the Chimera window and information about them is
shown in the SSD Browser:
The information includes PDB identifiers, checkboxes that toggle model display,
and menus for changing model color. N is the number of alpha-carbon
pairs, or matches, in the superposition, RMSD
is the root-mean-square deviation of the sets of alpha-carbons matched, and
Long Dist is the longest distance between matched alpha-carbons.
Ger-Lev is the Levitt-Gerstein probability score, described in:
"A unified statistical framework for sequence
comparison and structure comparison,"
M. Levitt and M. Gerstein, Proc. Natl. Acad. Sci. USA,
95,5913 (1998).
The -log(probability) is given, where probability represents the
likelihood that such a favorable superposition could be attained by chance
(proteins not structurally related);
larger values of -log(probability) indicate greater significance.
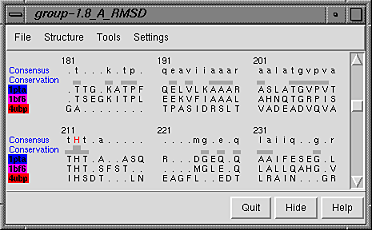
Both superpositions have an RMSD of 1.8 A because this was the selection
criterion; the settings shown in the
N vs N superpositions dialog figure were used to generate
this multiple superposition.
Clicking one of the buttons to the left of a PDB identifier opens
the pairwise superpositions
of that structure with the template structure.
Clicking Sequence Align opens a
structure-based sequence alignment corresponding to the
current superpositions, using the Chimera extension
Multalign Viewer.
In the structure-based sequence alignment, only the residues matched in
the superpositions are aligned in the sequence; unmatched residues are treated
as insertions (gaps in the other sequences).
The name of each sequence is shown in bold
over a rectangle matching the model color. This indicates
the sequence is associated with the structure shown in the
Chimera window.
CHIMERA TIPS
For new users,
here are a few basic tips
on moving structures, showing the command line,
and displaying ribbons in Chimera.
The Chimera User's Guide is a comprehensive manual in HTML format
(viewed with a web browser) that comes with your Chimera download.
Along with manual pages for each tool and command,
it contains tutorials for beginning and intermediate users.
Help... User's Guide in the Chimera menu opens the copy
that came with your local installation. This copy is the most appropriate
to use since it will match your installed Chimera software.
Structure Superposition Database / January 2007