Most images are shown at 1/3 size below. Use browser “save link as”
to get PNG files of images 1200px wide with transparent background.
Elaine C. Meng / meng@cgl.ucsf.edu /
home page
Post-Translational Modification
Luckily I had already done a lot of legwork to make
Fig 4 and related
morph movie for our
MCP paper. These illustrated not only the
difference between the unphosphorylated and phosphorylated (active)
conformations of FGFR1 kinase domain, but also the position of a
particular mutation. Since the latter is not necessary here, I removed
that additional complication from the image and morph movie and after a
few additional improvements in the script, remade the movie at high quality
in H.264 format. Still, they are similar enough to the published versions
that it may be prudent to include this citation,
as per journal instructions:
This research was originally published in Molecular & Cellular Proteomics.
JH Morris, EC Meng, TE Ferrin. Computational Tools for the
Interactive Exploration of Proteomic and Structural Data.
Molecular & Cellular Proteomics. 2010; 9:1703-1715.
© the American Society for Biochemistry and Molecular Biology.
(Such a citation is also needed for illustration #21 in the
previous set, but small font should be fine.)
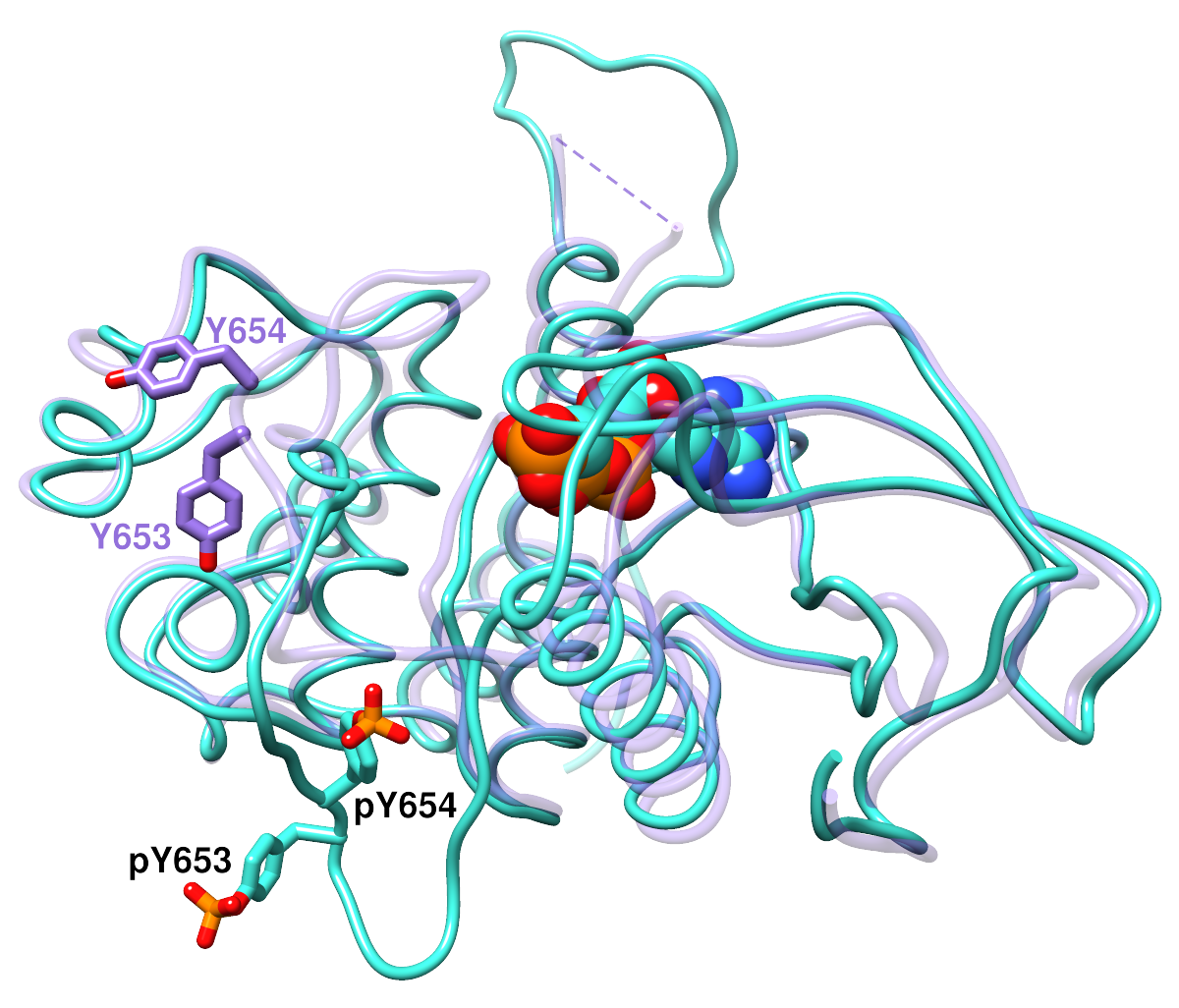 |
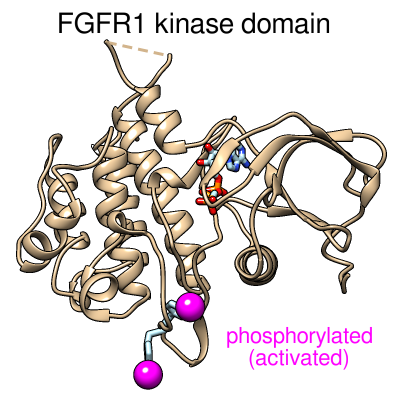
Superimposed structures of the basic fibroblast growth factor receptor 1
(FGFR1) kinase domain.
The backbones are shown as “licorice” ribbons,
and the side chains of interest are shown as sticks. pY, phosphotyrosine.
Heteroatoms are colored by element: oxygen, red; nitrogen, blue;
phosphorus, orange. The activated structure (PDB code
3GQI, chain A) with phosphorylated tyrosines
is shown in turquoise with black labels.
The activated structure also includes an ATP analog, displayed here
in the space-filling representation. The inactive conformation (PDB code
3C4F, chain A) is shown with a transparent purple
backbone and purple labels. (The purple dashed line indicates
structure information missing from the PDB file.)
|
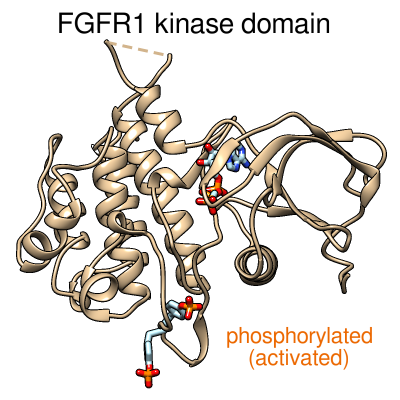
|
Morphing between the inactive and activated conformations of
the FGFR1 kinase domain.
Morphing between the same two structures as in the image above.
The protein backbone is shown as a tan ribbon.
The tyrosine side chains and ATP analog are shown as sticks,
with carbon in light blue, oxygen in red, nitrogen in blue,
and phosphorus in orange.
The ATP analog is faded in during the morph,
since it only fits in the activated conformation.
The image on the left is just a “still” from the movie.
[movie file H.264 400x400]
|
|
Same as above except with the phosphorylations simplified to magenta spheres.
[movie file H.264 400x400]
|
Mandelate Racemase
As suggested (it's as pretty as any enolase), here are some images of
mandelate racemase
(PDB 1MDR). Ribbon protein backbone,
sticks for the ligand (substrate analog (S)-atrolactate)
and the side chains of functional residues (as defined for the
mandelate racemase family in the SFLD).
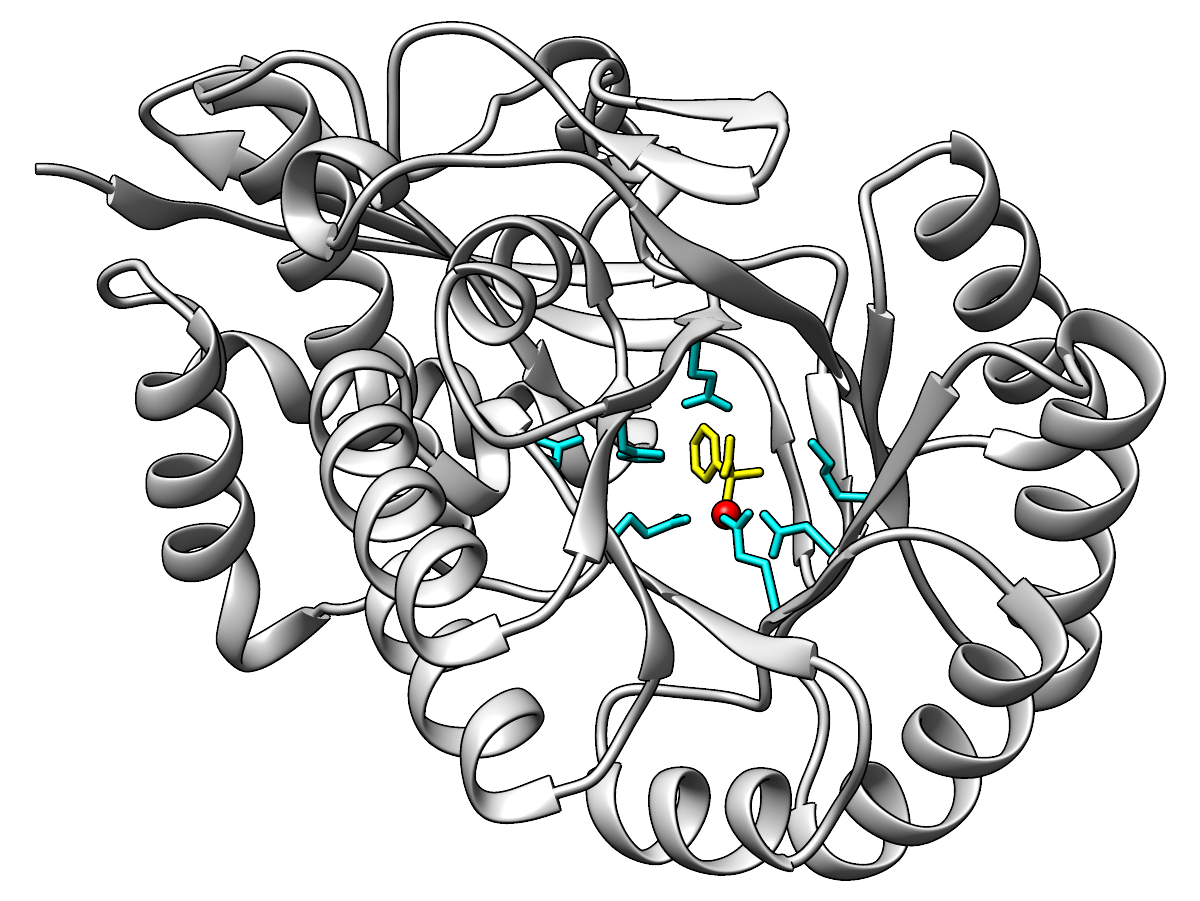 |
Ribbon light gray, functional residue sticks cyan, metal ion red,
ligand yellow.
|
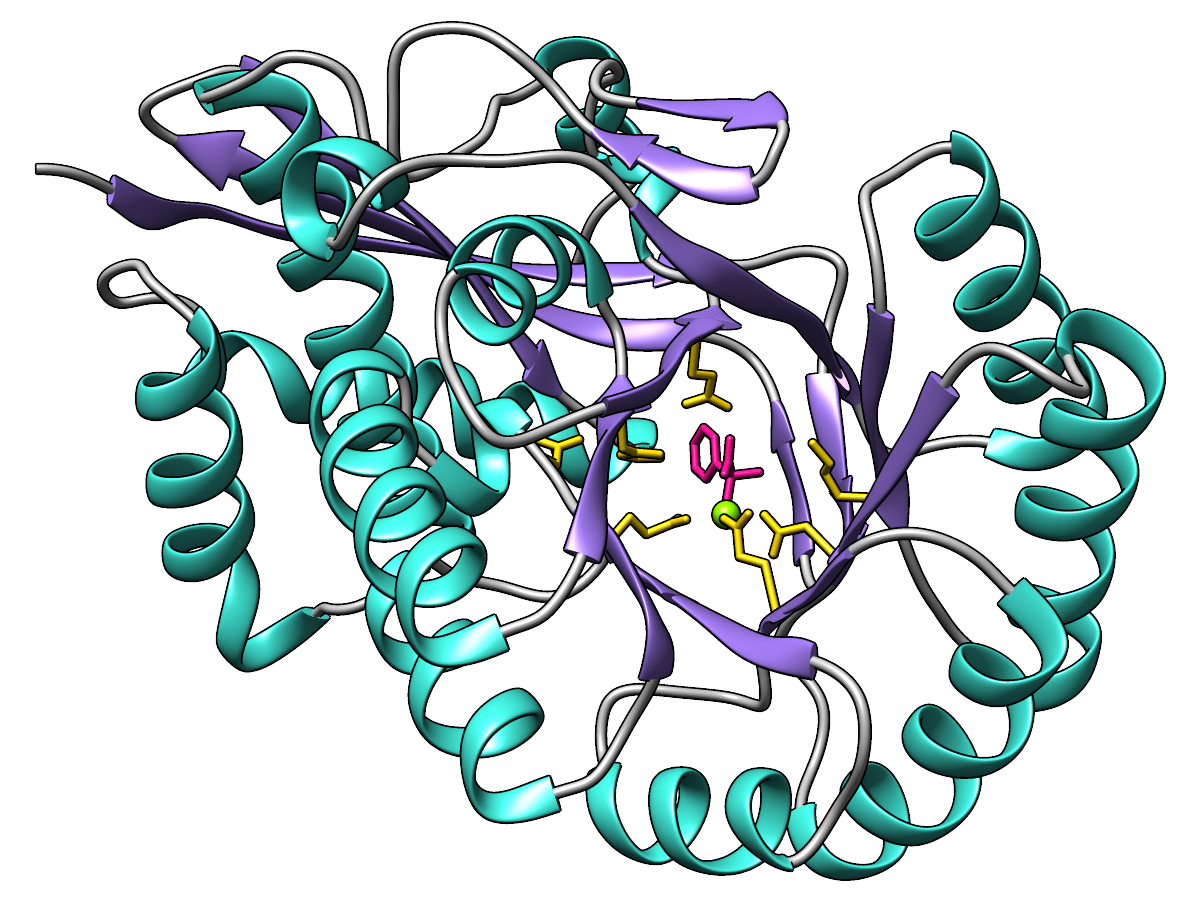 |
Ribbon colored by secondary structure:
α-helices in turquoise, β-strands in medium purple,
and the remainder (sometimes referred to as “coil”) in gray.
Functional residues gold, metal ion chartreuse, ligand in deep pink.
(Feel free to use different words to describe the colors!)
|
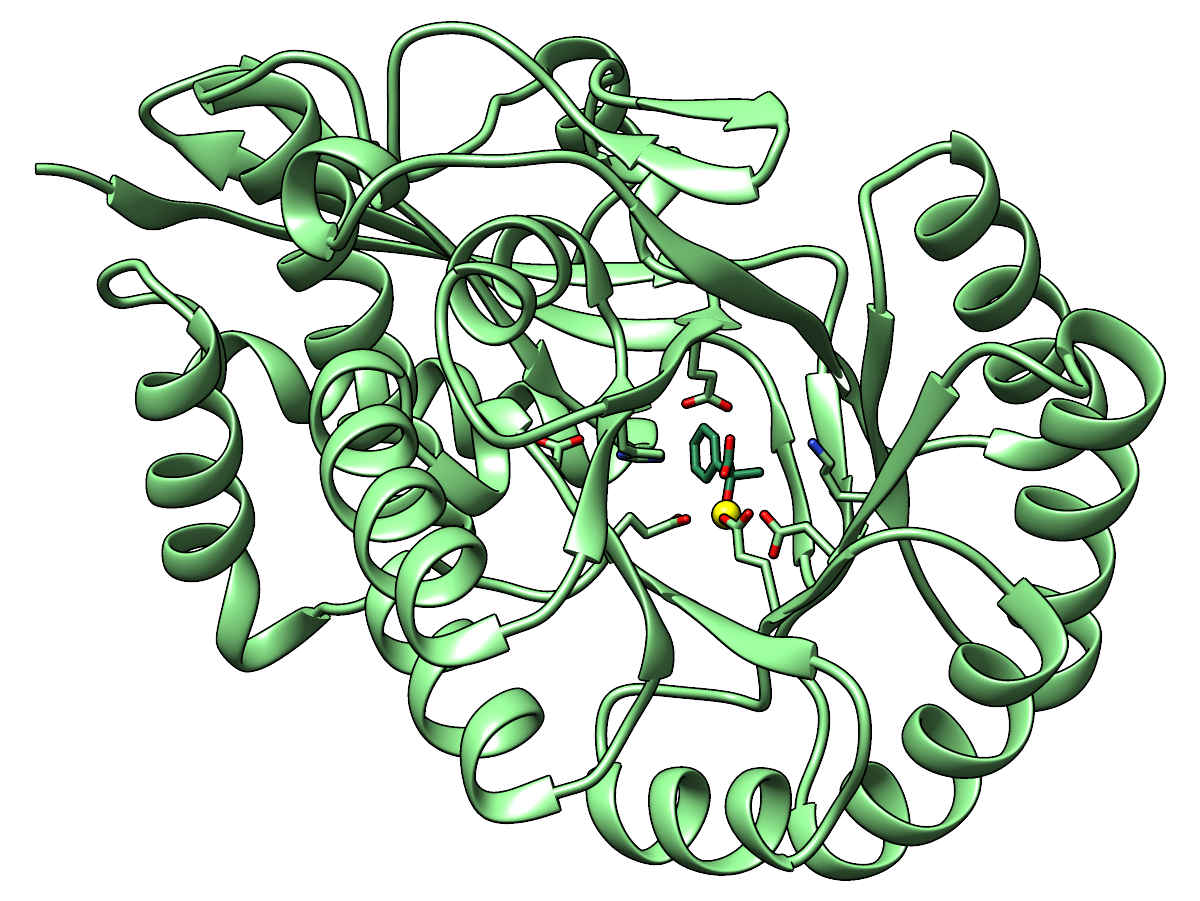 |
Ribbon and carbon atoms of the enzyme are light green, carbon atoms of the
ligand sea green. Oxygens are red, nitrogens blue, metal ion yellow.
|
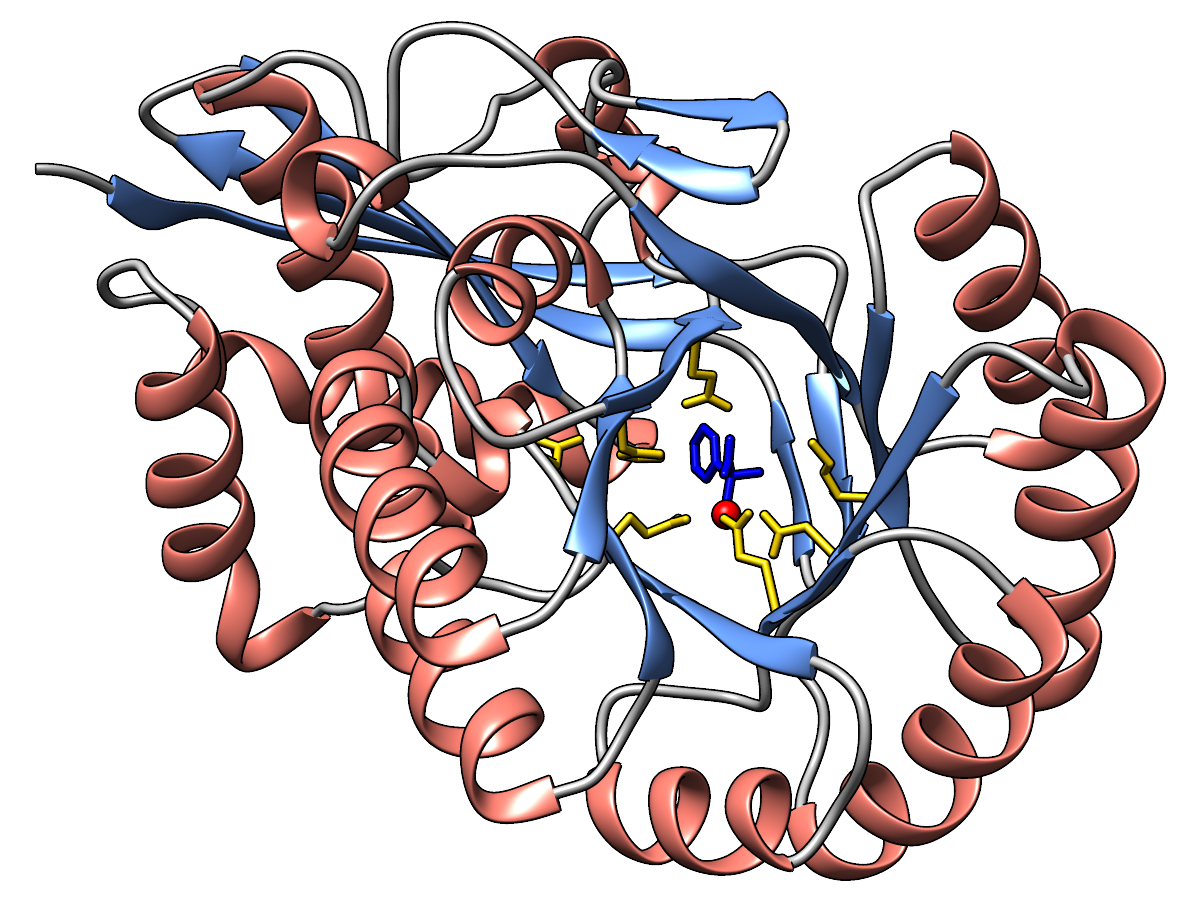 |
Ribbon colored by secondary structure:
α-helices in salmon, β-strands in cornflower blue,
and the remainder in gray.
Functional residues gold, metal ion red, ligand in blue.
|
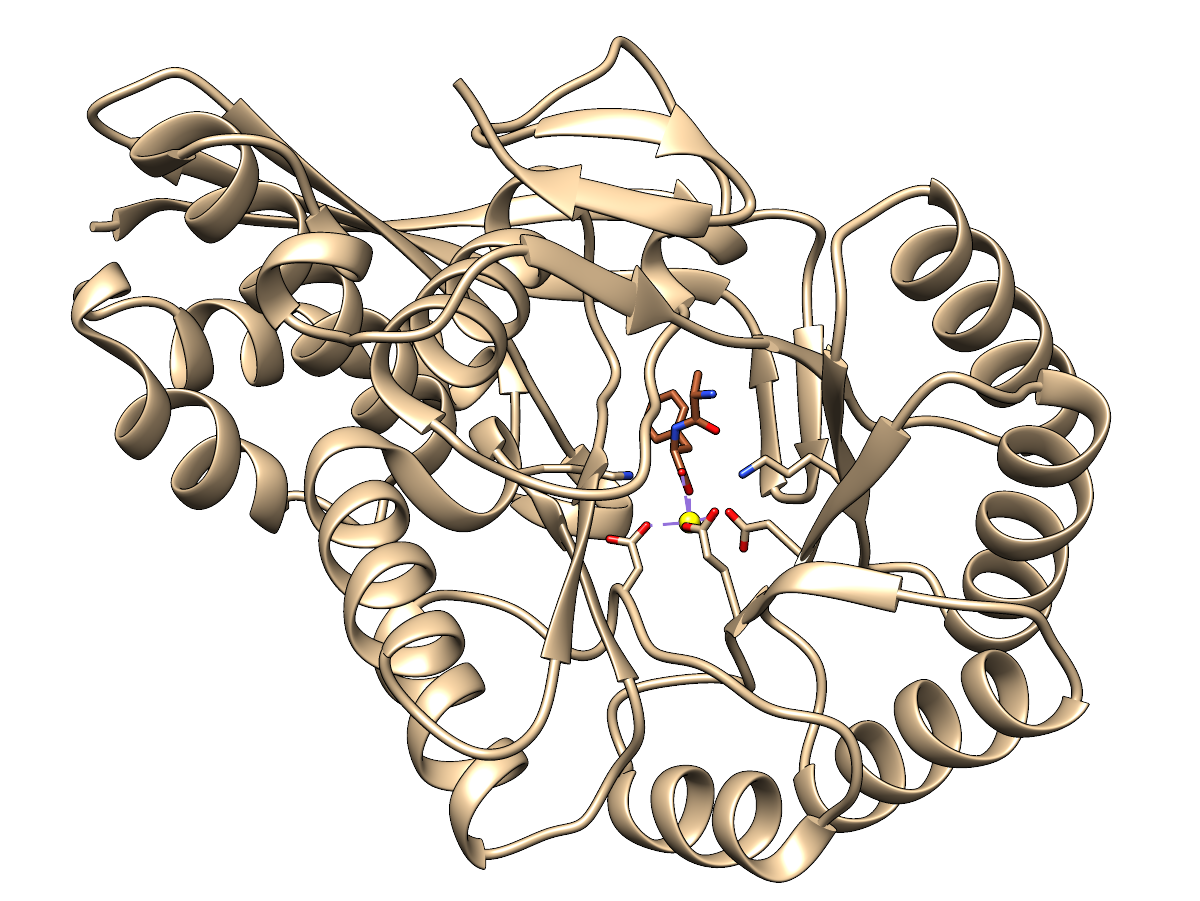
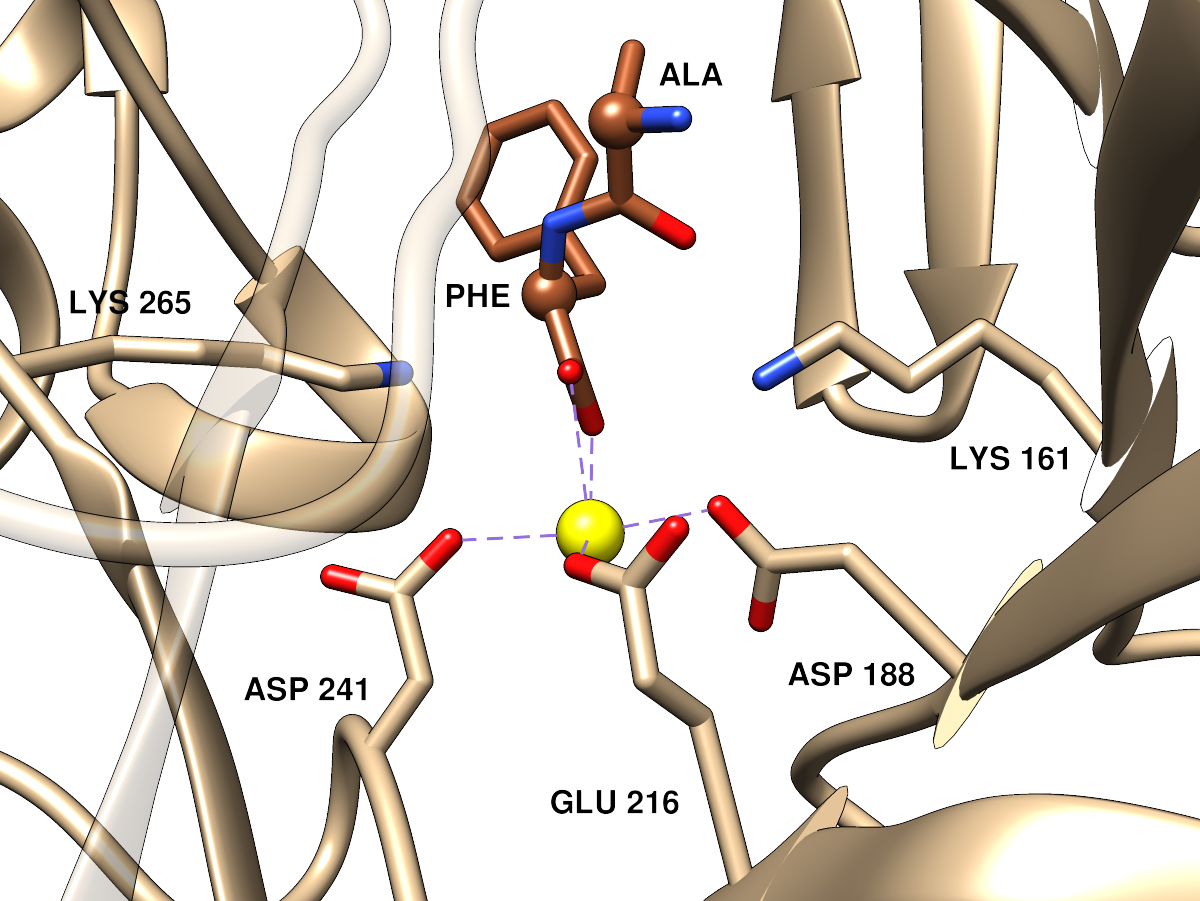
Dipeptide Epimerase
PDB 3JW7,
a dipeptide epimerase from Enterococcus faecalis
complexed with dipeptide L-Ile-L-Tyr.
This protein was characterized in:
Homology models guide discovery of diverse enzyme specificities among
dipeptide epimerases in the enolase superfamily.
Lukk T, Sakai A, Kalyanaraman C, Brown SD, Imker HJ, Song L, Fedorov AA, Fedorov EV, Toro R, Hillerich B, Seidel R, Patskovsky Y, Vetting MW, Nair SK, Babbitt PC, Almo SC, Gerlt JA, Jacobson MP.
Proc Natl Acad Sci USA. 2012 Mar 13;109(11):4122-7.
The sidechains of functional residues as defined in the SFLD
dipeptide
epimerase family page are shown, as well as the active site metal ion.
Ribbon and carbon atoms of the enzyme are tan, carbon atoms of the
ligand sienna. Oxygens are red, nitrogens blue, metal ion yellow.
There are purple dashes showing the coordination of the metal ion by
nearby oxygen atoms (but I can easily hide those if you would prefer).
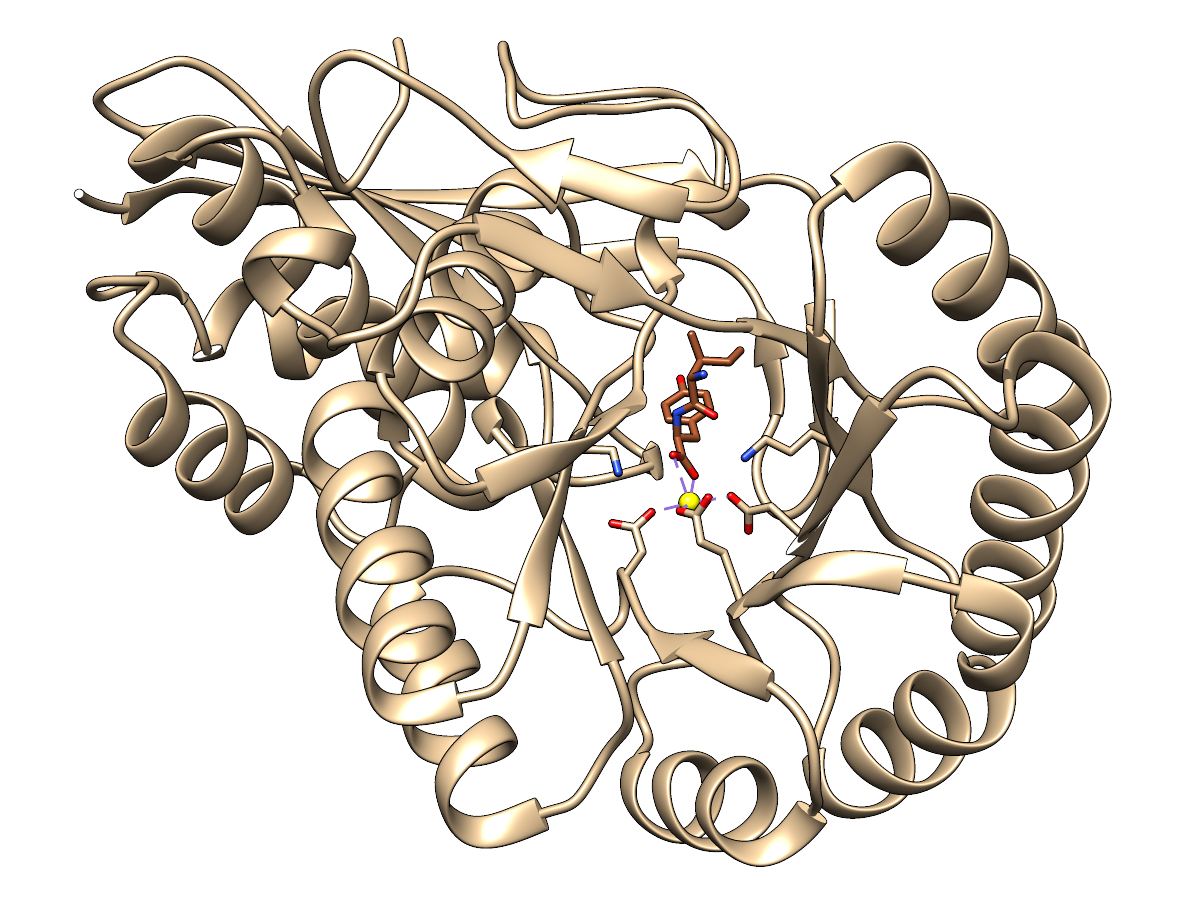 |
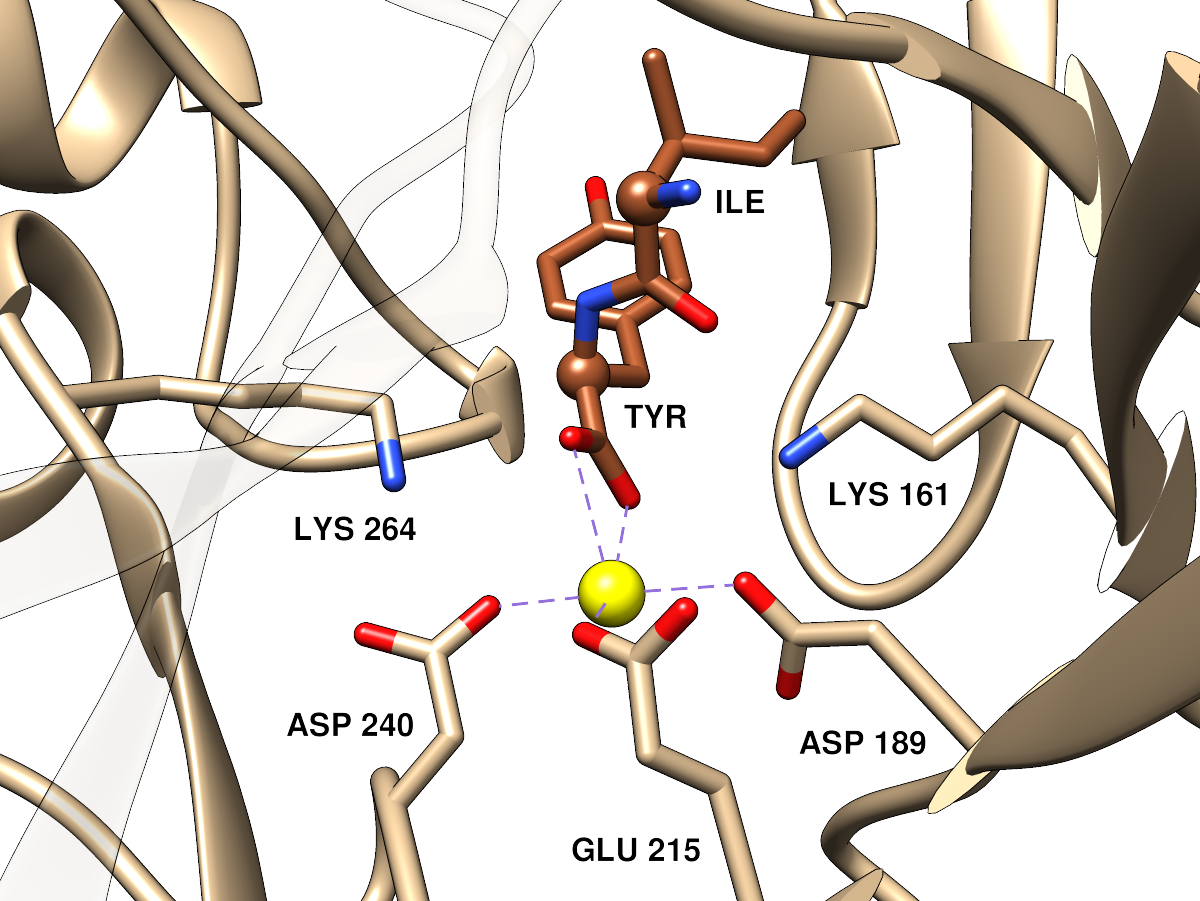 |
Approximately the same orientation and viewpoint as the far left image, but
zoomed in. Parts of the ribbon that would somewhat obscure the view have
been made transparent. The dipeptide ligand α-carbons are shown as balls.
(P.S. this is viewed from the opposite side as compared to SFLD active site
images for the enolase superfamily, but I thought it would be better to
match the orientation of the overall protein image)
|
Analogous images for
PDB 3DES: a dipeptide epimerase from Thermatoga maritima
complexed with dipeptide L-Ala-L-Phe.
This protein was characterized in:
Discovery of a dipeptide epimerase enzymatic function guided by
homology modeling and virtual screening.
Kalyanaraman C, Imker HJ, Fedorov AA, Fedorov EV, Glasner ME, Babbitt PC, Almo SC, Gerlt JA, Jacobson MP.
Structure. 2008 Nov 12;16(11):1668-77.