Extending CryoDRGN to Electron Tomography
CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Zhong ED, Bepler T, Berger B, Davis JH. Nat Methods. 2021 Feb;18(2):176-185.
[open source] [previous presentation]
Learning structural heterogeneity from cryo-electron sub-tomograms with tomoDRGN. Powell BM, Davis JH. Nat Methods. 2024 Aug;21(8):1525-1536. [tomodrgn github]
CryoDRGN-ET: deep reconstructing generative networks for visualizing dynamic biomolecules inside cells. Rangan R, Feathers R, Khavnekar S, Lerer A, Johnston JD, Kelley R, Obr M, Kotecha A, Zhong ED. Nat Methods. 2024 Aug;21(8):1537-1545. [cryodrgn github] (version 3.0.0-beta)
[back to paper list]
Cryo-EM:
- thousands to millions of particles of a target macromolecular complex are imaged, each from an unknown projection angle
- single-particle analysis (SPA) estimates the projection angle and the shape of each particle
- standard approaches required classification and separate treatment of the different classes of particles to avoid dealing with heterogeneity
- recently developed methods automatically handle heterogeneity of the particles to various extents, notably cryoDRGN, which can discern continuous conformational changes as well as distinct conformational and compositional differences
Cryo-ET:
- a single sample is imaged from a series of known tilt angles
- allows studying tissues, cells, and subcellular components down to the level of macromolecules in situ rather than requiring prior isolation
- sub-tomogram averaging (STA) is analogous to SPA in that many smaller volumes are identified as copies of the particle of interest, picked out, and combined to figure out its structure
- there have been some attempts to model heterogeneity of the picked particles in STA, but not to the extent of cryoDRGN for EM, until the works highlighted in this presentation...
CryoDRGN (Deep Reconstructing Generative Networks)
- unsupervised learning by two neural networks, encoder and decoder
- encodes 2D images into a latent space from which 3D maps are reconstructed
- assumes structures fall along a low-dimensional manifold in the latent space
- latent-space encoding
can be embedded in 2D for visualization, e.g. with PCA or UMAP)
- density maps can be generated from arbitrary values of the latent variable
TomoDRGN
- a generating tomoDRGN input from ET:
instances of the particle are picked, iterative STA generates a consensus map
for that type of particle, per-particle tilt images are re-extracted
from source data and annotated with parameters from STA (pose, defocus, etc.)
- b tomoDRGN:
each particle's tilt images are independently passed through encoder A and
then jointly passed through encoder B, resulting in a latent space from
which 3D maps can be reconstructed by the decoder
- c: symbols that will be used in subsequent figures to clarify
how volumes were generated (VAE = variational autoencoder)
- The decoder network can be used alone (without latent space learning)
for homogeneous reconstruction.
- CryoDRGN maps individual images to the latent space, but is not
constrained to map differentially tilted images of the same particle
to consistent regions in that space; thus tomoDRGN has encoder A
(analogous to the cryoDRGN encoder) + encoder B to integrate these intermediate
embeddings into a single latent embedding per particle.
- Standard VAE training minimizes loss between input and reconstructed output.
Additional STA-Specific Approaches
- model signal-to-noise ratio (SNR) as a function of tilt angle and
accumulated electron dose (downweight highly tilted and radiation-damaged
images, especially at high frequencies)
- apply optimal dose filtering to Fourier coordinates evaluated by the
decoder (mask frequencies for which the dose was suboptimal, thus
limiting the computational burden of training with little effect on accuracy)
- account for missing tilt images by finding the particle with the fewest tilt images and using only that number (randomly sampled) for the others; this permutes tilt images at random and forces encoder B to learn a permutation-invariant mapping of the particle to latent space
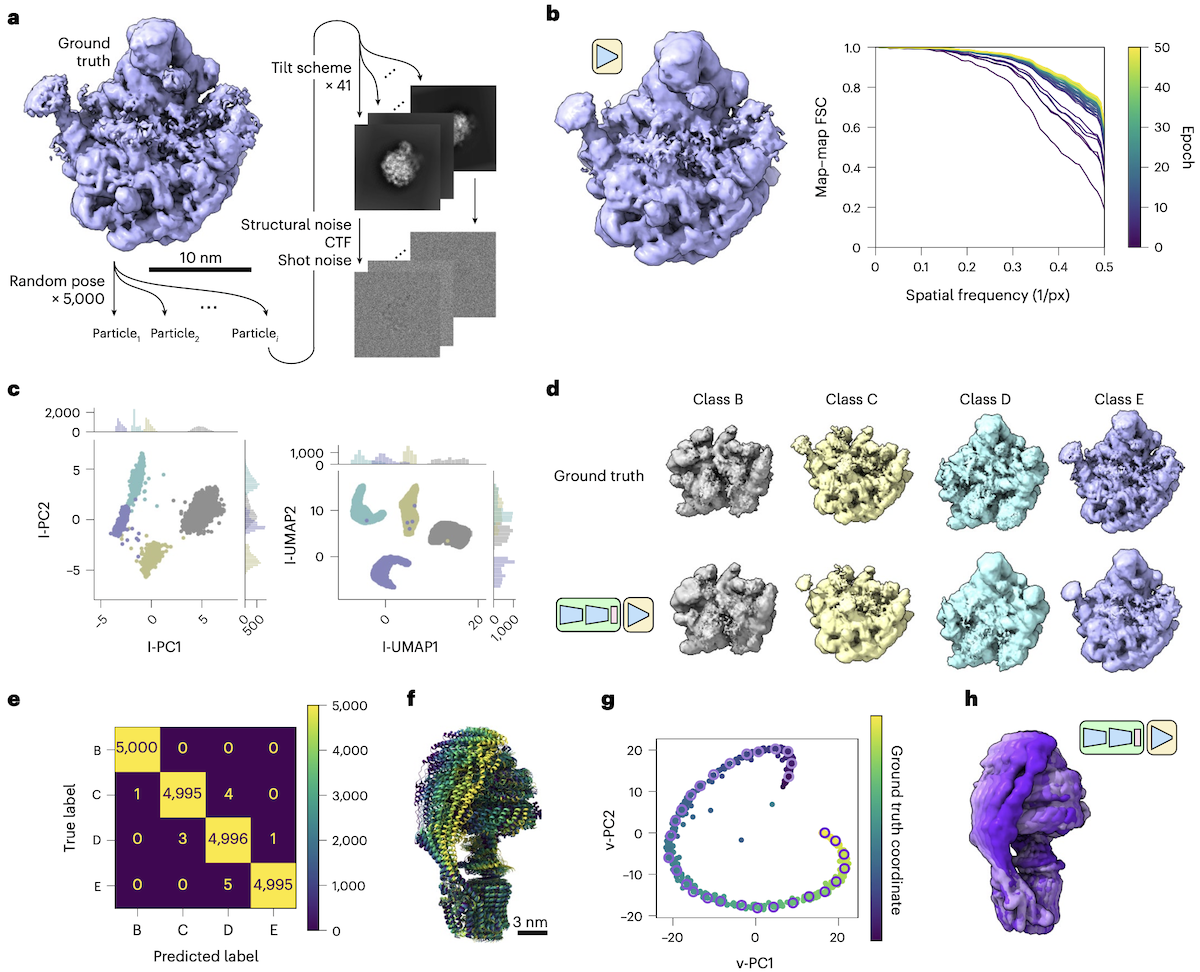
TomoDRGN recovers compositional and conformational heterogeneity in simulated data
- a: four assembly states of the ribosomal large subunit
(see d), 5000 particles each, 41 tilt images
- b: tomoDRGN homogeneous reconstruction of class E data
converges rapidly. Also noted (supp data, not shown here):
- weighting and filtering each improved resolution
- filtering significantly ↓ runtime
- random tilt sampling ↑ correlation with ground truth
- c: latent encoding for the combined dataset of four classes
colored by k-means class (k=4) after 24 epochs
- d: ground truth volumes vs. tomoDRGN reconstructions from median
latent encoding of each k-means class
- e: confusion matrix of ground truth labels vs. k-means class labels
- f: ATP synthase conformations along simulated reaction coordinate
- g,h: TomoDRGN latent-space embedding and reconstructed maps show a smooth and continuous trajectory
CryoDRGN fails to consistently encode structural heterogeneity in simulated tilt series
- (same simulated tilt series of four states of the ribosomal large subunit)
- a: architecture schematics
- b: UMAP of final epoch latent space embeddings
- c: UMAP with k-means latent space classification (k=4)
- d: confusion matrix of ground truth labels vs. k-means class labels
- e: reconstructed maps for k-means cluster centers marked in panel c
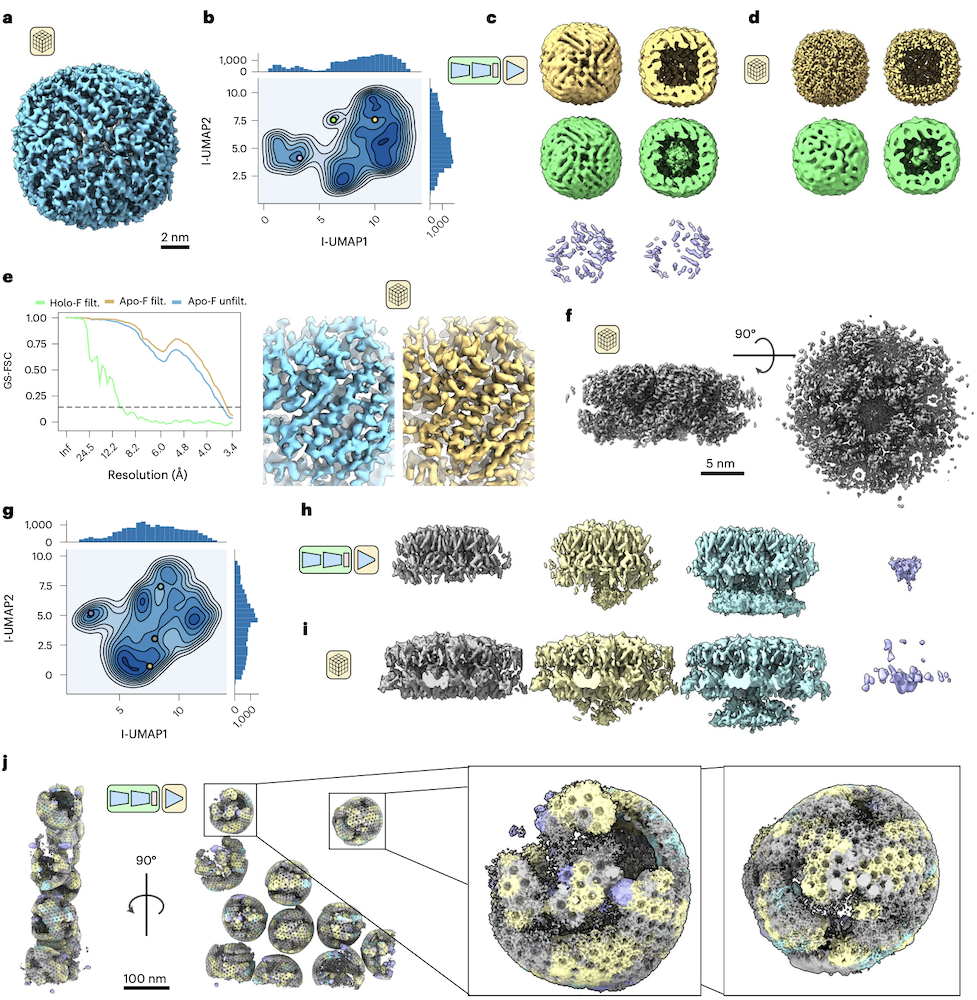
TomoDRGN finds heterogeneity among primarily homogeneous purified particles
- a: consensus STA apoferritin refined with C1 symmetry (EMPIAR-10491, n=25,381 particles)
- b: tomoDRGRN latent encoding
- c: volumes from the 3 encodings marked in b: apoferritin (yellow, ~65%), holoferritin (green, ~2%), uniterpretable (~33%, likely from errors in particle picking)
- d: separate STA reconstructions for tomoDRGN-classified apo- and holoferritin
- e: tomoDRGN filtering improves STA result
- f: consensus STA HIV Gag refined with C1 symmetry (EMPIAR-10164, n=18,325 particles)
- g: tomoDRGRN latent encoding
- h: volumes from the 4 encodings marked in g
- i: weighted back-projection of separate tomoDRGN classes
- j: EMPIAR-10164 tomogram reconstructed with tomoDRGN; Gag hexamer volumes colored as above, showing clustering of hexamers in classes with greater nucleocapsid density (lower parts in h,i)
TomoDRGN resolves high-resolution features in subtomograms collected in situ
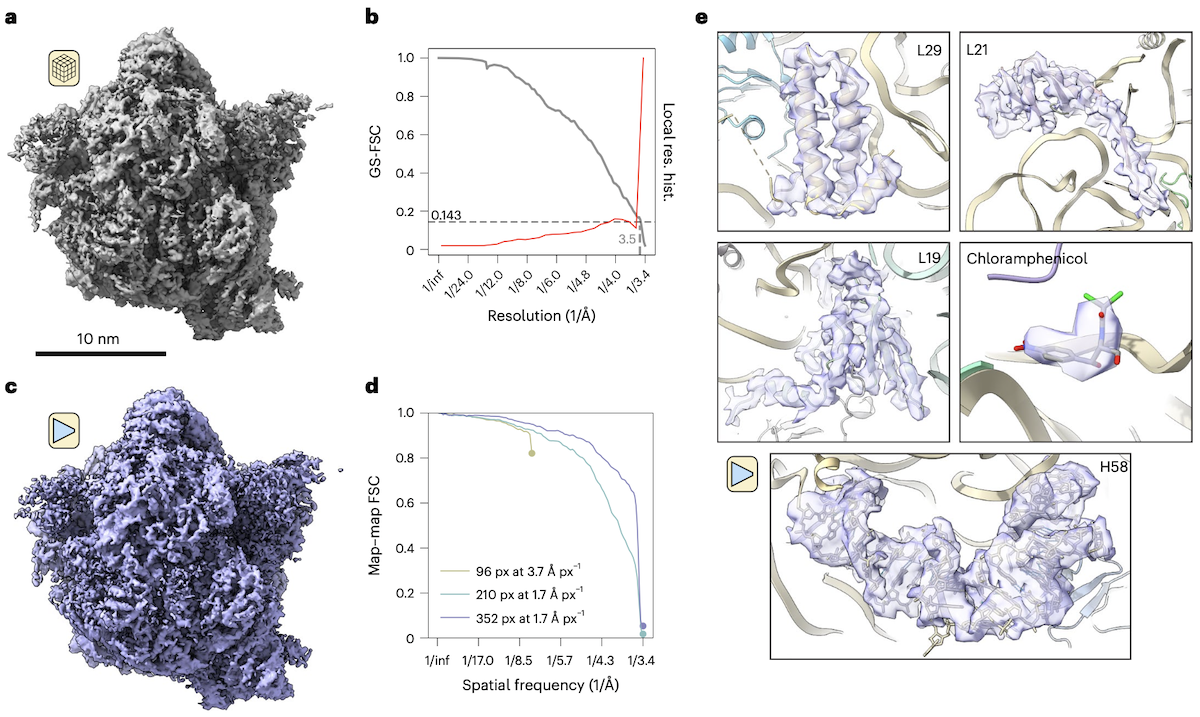
- a: STA reconstruction of M. pneumoniae 70S ribosome (EMPIAR-10499, n=22,291 particles)
- b: gold standard FSC curve between STA half-maps (gray), local resolution (red)
- c: tomoDRGN homogeneous reconstruction (decoder module only)
- d: map-map FSCs of tomoDRGN reconstructions (different box & pixel sizes) vs. the corresponding STA volumes
- e: closeups from the reconstruction shown in c (tomoDRGN recapitulates features observed in STA)
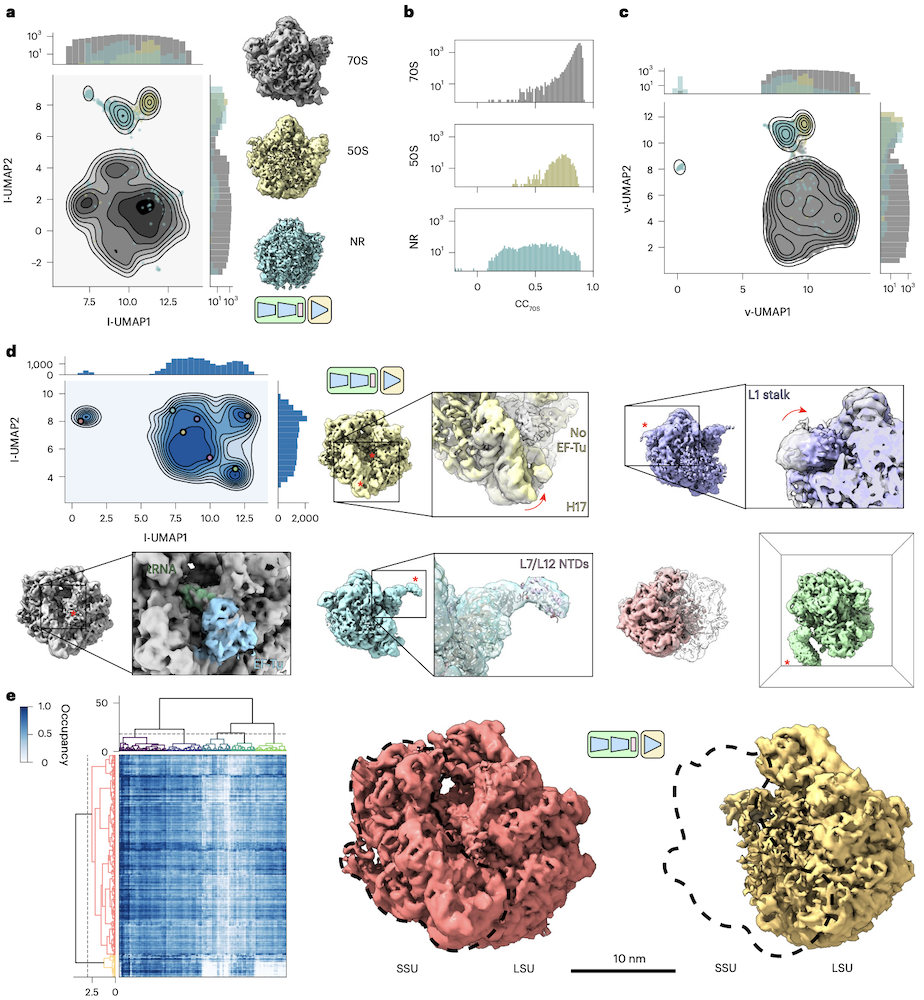
TomoDRGN finds structural heterogeneity in ribosomes imaged in situ
- a: heterogeneous tomoDRGN trained on a downsampled
version of the same data (EMPIAR-10499, n=22,291)
resolves 70S (whole ribosome), 50S (large subunit only), NR (nonribosomal)
- b: cross-correlation values of map from each particle's latent
embedding vs. the STA map
- c: UMAP of first 128 principal components of volumes from b
- d: tomoDRGN latent encoding of ribosomal particles only (n=20,981),
reconstructions for marked points highlighting variable regions
- e: MAVEn-clustered heatmap of 500 volumes sampled from the tomoDRGN model shows separation of 70S vs. 50S particles. MAVEn (Model-based Analysis of Volume Ensembles) is a coarse-grained approach to quantify occupancy and flexibility of diverse structural elements.
Examples of nonribosomal particles identified by tomoDRGN
- a: tomoDRGN latent encoding (same data as previous, n=22,291), eight
representative nonribosomal particles identified by finely clustering latent space
(k-means clustering with k=100)
- b: tomograms in slice view with nonribosomal particles marked
- c: RELION3 multiclass (k=5) ab initio subtomogram volume generation using particles annotated by tomoDRGN as nonribosomal (n=1310)
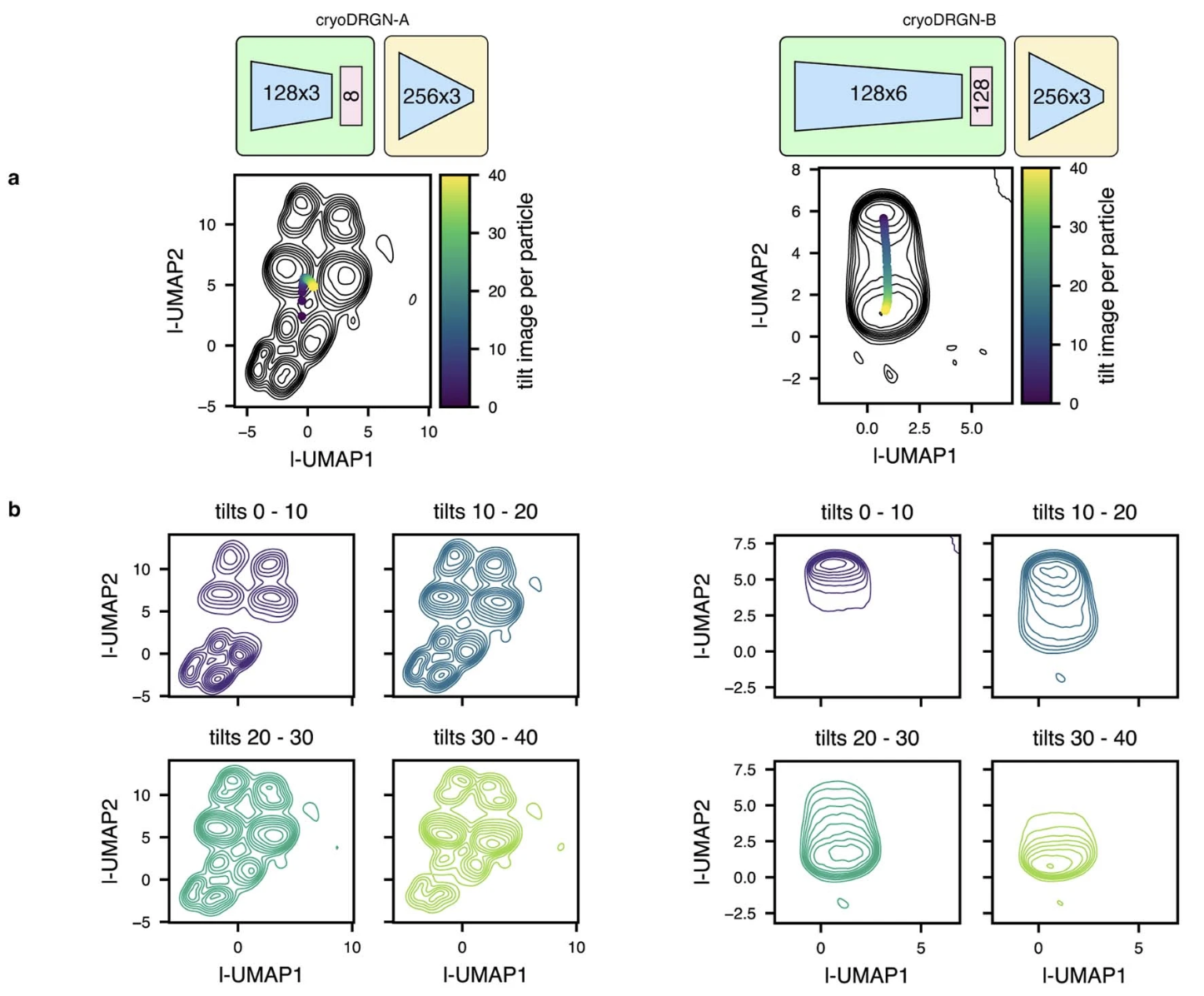
CryoDRGN learns errant heterogeneity in ET of ribosomes in situ
- a,b: two cryoDRGN models trained on the unfiltered particle stack
of ribosomes imaged in situ (same dataset as previous, 22,291 particles
treated as 913,931 images)
- In the lower left of each panel is a reference 70S ribosome volume sampled from the tomoDRGN model shown in the previous slide.
CryoDRGN latent space embeddings exhibit undesirable correlations with tilt image index
TomoDRGN captures compositional heterogeneity in situ
- a: latent encoding from "intermolecular tomoDRGN"
trained on less-cropped images to include ribosome binding partners
(n=20,981 re-extracted with box size ~3X the particle radius)
- b: violin and projection plots of the distance
from the class of particle to its nearest-neighbor ribosome
(red box is
typical inter-ribosome distance in prokaryotic polysomes)
- c: distribution of classes per tomogram; width proportional
to each tomogram's particle count
- d: screenshot from tomoDRGN's interactive viewer marking
ribosomal particles with cones, and
membrane-associated ones additionally with spheres
- e: latent encoding of membrane-associated ribosomes only (n=482)
- f: STA reconstruction of membrane-associated ribosomes with fitted structure of AlphaFold-predicted secDF (part of Sec translocon)
Other Findings and Discussion
- performance is best on large, abundant particles
- the approach is iterable, i.e. retraining can focus on
newly identified or more thoroughly "purified" classes
- performance is robust to encoder network architecture hyperparameters
(supp tables 2-4, not shown)
- larger decoder networks support learning of higher-resolution features as the expense of slower model training
(supp figs 1,2, not shown)
- even with random tilt sampling, mild overfitting is possible;
users are encouraged to guard against overfitting by using the
provided "analyze_convergence" tool at regular intervals
- mapping single sub-tomogram volumes to single latent coordinates could
theoretically be done within the cryoDRGN framework, but would likely be:
- less computationally tractable due to cubic scaling of voxels per particle
- predisposed toward learning heterogeneity driven by missing wedge artifacts
- potential limitations from tomoDRGN inputs, but that were not found
to cause significant problems:
- inaccuracies in pose estimation during upstream STA processing
- background signals superimposed on the particle at certain tilt angles
- symmetry relaxation or expansion of symmetric complexes is recommended
before tomoDRGN analysis because its decoder is best suited to particles
posed in C1 (no assumption of symmetry)
- for visualization in ChimeraX, tomoDRGN uses the subtomo2chimera script to place each particle’s volume at its source location and orientation in the tomogram context