CryoDRGN
CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Zhong ED, Bepler T, Berger B, Davis JH. Nat Methods. 2021 Feb;18(2):176-185.
- Cryo-EM single-particle analysis has proven powerful in determining
the structures of rigid macromolecules. However, many complexes
exhibit conformational and compositional heterogeneity that pose
a major challenge to existing 3D reconstruction methods.
- cryoDRGN uses deep neural networks to reconstruct continuous distributions
of 3D density maps reflecting the heterogeneity within single-particle
cryo-EM datasets.
- CryoDRGN is open-source software freely available from http://cryodrgn.csail.mit.edu
[back to paper list]
Existing Methods: Homogeneous vs. Heterogeneous
- Homogeneous reconstruction considers the data to represent a single conformational (and compositional) state
- Many methods require classifying particles into quasi-homogeneous subsets, then using the different classes for separate reconstructions
Problems with this approach:
- The number of states/classes is not known a priori and may be difficult to determine accurately
- Using a discrete number of classes does not account for continuous variations in conformation
More-Advanced Heterogeneous Methods
- RELION multibody refinement
– models the structure as the sum of user-defined rigid bodies
that can rotate relative to one another
- cryoSPARC 3D variability analysis
– PCA-based, represents variability within a linear subspace
(which may be misleading when structural deformations are not
well described by linear interpolations between basis vectors)
- manifold embedding
– binning particles along the data manifold followed by
traditional homogeneous reconstruction
- others...
CryoDRGN (Deep Reconstructing Generative Networks) Method
- unsupervised learning by two neural networks, encoder and decoder
- encodes 2D images into a latent space from which 3D maps are reconstructed
- assumes structures fall along a low-dimensional manifold in the latent space
- dimensionality specified by the user, 10 dimensions generally effective
- density maps can be generated from arbitrary values of the latent variable
- regions of latent space rich in impurities or artifacts can be filtered out
- initial filtering run can be done on downsampled data (Fourier clipping)
What does CryoDRGN allow researchers to do?
- get latent-space encoding of particle distribution
(which can be embedded in 2D for visualization, e.g. with PCA or UMAP)
- generate density maps for exploratory analysis
- extract particle subsets for use with other tools
- generate trajectories of molecular motions (as maps)
- mainly, to characterize cryoEM dataset heterogeneity and generate maps of the various states without having to classify the particles ahead of time
“Homogeneous” High-Resolution Data
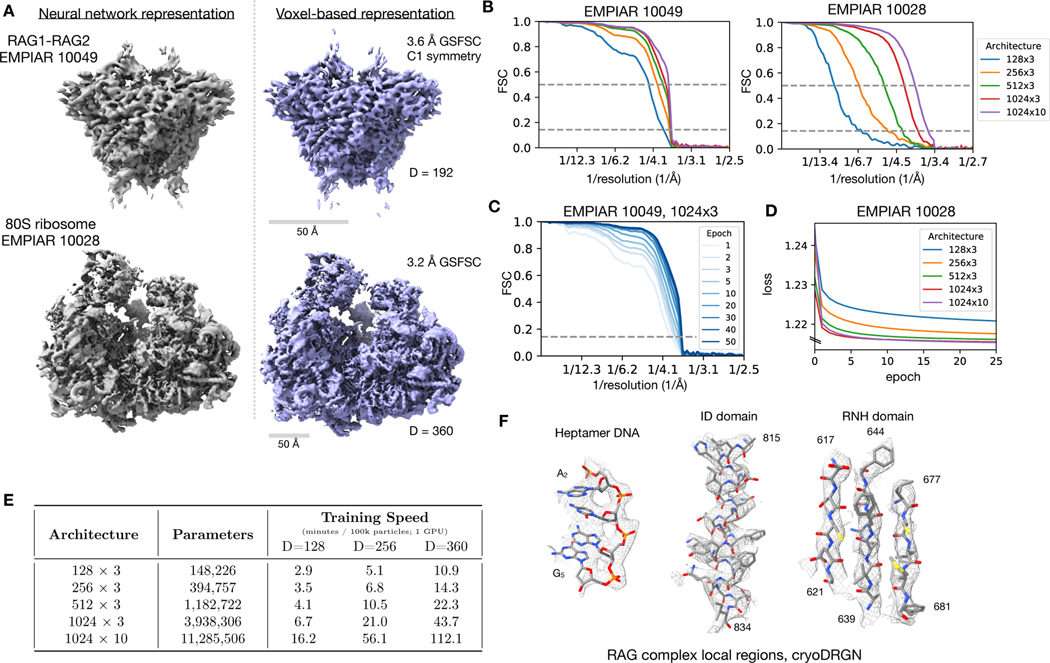
EMPIAR (Electron Microscopy Public Image ARchive) – raw data for some EMDB entries. 2022-08-02: 975 datasets, 2.0 PB. For RAG1-RAG2 (EMPIAR 10049), P. falciparum 80S ribosome (EMPIAR 10028):
- A. cryoDRGN (1024 x 3, "no latent variable input") vs. traditional (cryoSPARC)
- B. FSC (Fourier shell correlation) curves for NN architectures (E), resln <4Å at FSC 0.5
- C. FSC evolution over training epochs, D. similar for loss
- E. Architecture is tradeoff between resolution and speed
- F. CryoDRGN map with RAG1-RAG2 atomic structure (PDB 3jbx)
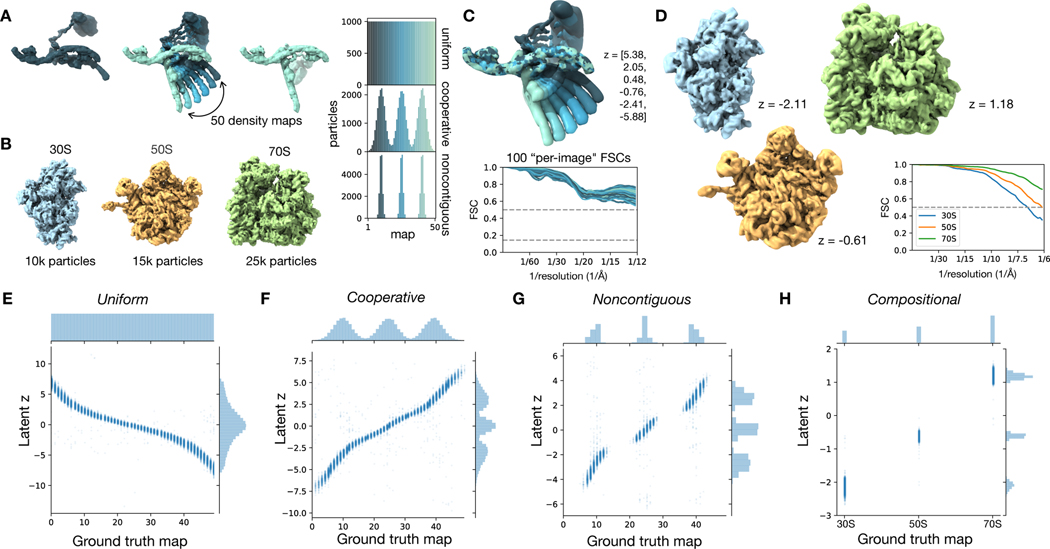
Simulated Heterogeneity
- A. Conformational: Uniform sampling along 1D reaction coordinate of rotating a single dihedral, Cooperative biased to specific states, Noncontiguous with unobserved transition states
- B. Compositional from mixing bacterial 30S, 50S, and 70S ribosomal complexes
- C. for the Uniform data, sample of cryoDRGN maps and per-image FSCs (output maps vs. ground-truth maps across the reaction coordinate)
- D. CryoDRGN maps from the Compositional data
- E-H. Latent-space encodings (using 1D model) consistent with ground truth
Heterogeneity in “Homogeneous” Data
- A. RAG1-RAG2 maps from original authors based on 2 particle classes: signal end (only core resolved) and paired (other parts visible)
- B,C. In the signal end data, cryoDGRN (10D) finds positions of formerly unresolved parts (ribbons show the previous paired structure for comparison) possibly important for function [video]
- D,E. In the Pf80S ribosome data, cryoDRGN (10D) identifies a rotated-subunit substate
Highly Heterogeneous Data
- – E. coli large ribosomal subunit (50S) undergoing assembly (EMPIAR-10076)
- – Original analysis found 13 classes grouped into 4 major assembly states
- A-D. CryoDRGN 1D and 10D latent space representations and filtering (details in suppl fig 5)
- E-G. CryoDRGN finds the 4 states without prior classification (dashed outline shows mature subunit)
- F,G. Latent space UMAP colored/labeled by states from original analysis
- H. Some particles ("A" in panel G) are whole ribosome (70S = 30S + 50S)
- I. C4 is previously unreported assembly intermediate
Large Continuous Conformational Changes
- – Pre-catalytic spliceosome (EMPIAR-10180)
- – Original expert classification suggested a continuum of conformations with large motions of the SF3b subcomplex
- A,B. CryoDRGN (10D) finds several clusters; filter to remove those with likely imaging artifacts (iii), missing the SF3b subcomplex (iv), or that have the additional U2 core (v)
- C,D. Maps generated along principal components show large movements of the SF3B subcomplex relative to the rest of the structure [video]
Others Can Use CryoDRGN
-
Conformational landscape of the yeast SAGA complex as revealed by cryo-EM.
Vasyliuk et al., Sci Rep. 2022 Jul 19;12(1):12306.
-
Structural basis of human telomerase recruitment by TPP1-POT1.
Sekne et al., Science. 2022 Mar 11;375(6585):1173-1176.
-
A late-stage assembly checkpoint of the human mitochondrial ribosome large subunit.
Rebelo-Guiomar et al., Nat Commun. 2022 Feb 17;13(1):929.
-
Catalytic trajectory of a dimeric nonribosomal peptide synthetase subunit with an inserted epimerase domain.
Wang et al., Nat Commun. 2022 Feb 1;13(1):592.
- Structure and dynamics of the chromatin remodeler ALC1 bound to a PARylated nucleosome. Bacic et al., eLife. 2021 Sep 6;10:e71420.
Can We Use CryoDRGN in ChimeraX?
- ...or its outputs after training?
- related ticket #6106: Efficiently select maps by visual inspection