 |
The primary use of sym is to facilitate symmetrical placement of copies of a structure within related volume data, usually a density map. The symmetry of the data can be specified in either of two ways:
The keyword contact indicates that only copies with any atom within contact-dist of the original molecule should be generated. The keyword range indicates that only copies with centers within range-dist of the center of the original molecule should be generated. A structure's center is defined as the center of its bounding box. It may be necessary to use one of these options or simply to specify fewer matrices to avoid creating too many atoms.
A secondary use of sym is to generate BIOMT-described copies of a molecule model, where that model is specified as both molmodel and refmodel. Since a model cannot be moved relative to itself, the dynamic updating does not come into play in this situation.
The command ~sym removes the copies of molmodel generated with sym. If the volume refmodel is closed, the copies of molmodel will no longer update to preserve symmetry when the original molecule is moved.
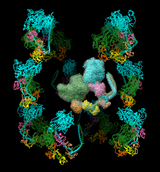
BIOMT matrices can be added to PDB files with a text editor. The image shows twelve copies of myosin arranged helically, as specified by the following twelve matrices for PDB entry 1i84 (the first is simply an identity matrix that does not specify an additional copy):
REMARK 350 BIOMOLECULE: 1 REMARK 350 APPLY THE FOLLOWING TO CHAINS: S, T, U, V, W, Z REMARK 350 BIOMT1 1 1 0 0 0 REMARK 350 BIOMT2 1 0 1 0 0 REMARK 350 BIOMT3 1 0 0 1 0 REMARK 350 BIOMT1 2 0 -1 0 0 REMARK 350 BIOMT2 2 1 0 0 0 REMARK 350 BIOMT3 2 0 0 1 0 REMARK 350 BIOMT1 3 -1 0 0 0 REMARK 350 BIOMT2 3 0 -1 0 0 REMARK 350 BIOMT3 3 0 0 1 0 REMARK 350 BIOMT1 4 0 1 0 0 REMARK 350 BIOMT2 4 -1 0 0 0 REMARK 350 BIOMT3 4 0 0 1 0 REMARK 350 BIOMT1 5 0.866025 -0.5 0 0 REMARK 350 BIOMT2 5 0.5 0.866025 0 0 REMARK 350 BIOMT3 5 0 0 1 145 REMARK 350 BIOMT1 6 -0.5 -0.866025 0 0 REMARK 350 BIOMT2 6 0.866025 -0.5 0 0 REMARK 350 BIOMT3 6 0 0 1 145 REMARK 350 BIOMT1 7 -0.866025 0.5 0 0 REMARK 350 BIOMT2 7 -0.5 -0.866025 0 0 REMARK 350 BIOMT3 7 0 0 1 145 REMARK 350 BIOMT1 8 0.5 0.866025 0 0 REMARK 350 BIOMT2 8 -0.866025 0.5 0 0 REMARK 350 BIOMT3 8 0 0 1 145 REMARK 350 BIOMT1 9 0.866025 0.5 0 0 REMARK 350 BIOMT2 9 -0.5 0.866025 0 0 REMARK 350 BIOMT3 9 0 0 1 -145 REMARK 350 BIOMT1 10 -0.5 0.866025 0 0 REMARK 350 BIOMT2 10 -0.866025 -0.5 0 0 REMARK 350 BIOMT3 10 0 0 1 -145 REMARK 350 BIOMT1 11 -0.866025 -0.5 0 0 REMARK 350 BIOMT2 11 0.5 -0.866025 0 0 REMARK 350 BIOMT3 11 0 0 1 -145 REMARK 350 BIOMT1 12 0.5 -0.866025 0 0 REMARK 350 BIOMT2 12 0.866025 0.5 0 0 REMARK 350 BIOMT3 12 0 0 1 -145There is a Quicktime movie of the myosin copies moving symmetrically at the Chimera Web site.
See also: matrixcopy, volume, Volume Viewer, Multiscale Models, Unit Cell