Unit Cell builds crystallographic unit cells using information in PDB files. A crystallographic unit cell consists of a unique set of coordinates, duplicated and transformed according to the crystallographic and (if present) noncrystallographic symmetries in the crystal. Unit Cell can be used to regenerate the full unit cell, or only those parts defined by crystallographic symmetry or noncrystallographic symmetry. See also: Multiscale Models, sym, fetching PQS files, Crystal Contacts
There are several ways to start Unit Cell, a tool in the Higher-Order Structure category.
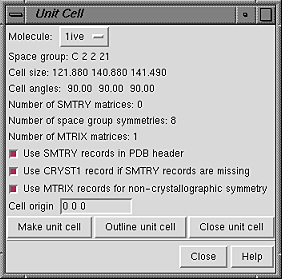 Once a PDB file has been
opened in Chimera, Unit Cell can generate symmetry-related
copies if the PDB header contains sufficient information.
In the dialog, Molecule can be set to any open molecule model.
The space group, unit cell parameters, and numbers of transformation matrices
available for the current Molecule are shown.
If the PDB file has no CRYST1 record, the space group and cell parameters
fields will be blank. If the PDB file also lacks SMTRY and MTRIX
information, or if the model was not read from a PDB file, it will not
be possible to build a unit cell.
Once a PDB file has been
opened in Chimera, Unit Cell can generate symmetry-related
copies if the PDB header contains sufficient information.
In the dialog, Molecule can be set to any open molecule model.
The space group, unit cell parameters, and numbers of transformation matrices
available for the current Molecule are shown.
If the PDB file has no CRYST1 record, the space group and cell parameters
fields will be blank. If the PDB file also lacks SMTRY and MTRIX
information, or if the model was not read from a PDB file, it will not
be possible to build a unit cell.
Make unit cell loads copies of the coordinates from the same source as the originally opened model (a local file or a file fetched from the web) and transforms them using the symmetry information specified by the checkbox settings. Pressing the button a second time will create a new set of copies without deleting the first set. Outline unit cell shows a white box outlining the unit cell; this outline can be hidden or closed with the Model Panel. Close unit cell deletes all open copies of the molecule, except for the original. All copies will be deleted, not just the ones specified by the current checkbox settings.
The unit cell can be shifted along its axes by changing the Cell origin coordinates and pressing Enter (return). This will shift the outline (if present) and may rearrange the copies within. Cell origin coordinates are in unit cell lengths and describe translations of the unit cell along its axes, which generally differ from the axes of the viewing coordinate system. Values of a coordinate that differ by integer amounts are equivalent; e.g., (0.3 0 0) is equivalent to (-0.7 0 0) and (4.3 0 0). The shift will occur in the direction that maintains the center of the original copy within the box. Shifting allows all contacts to be visualized even though only a single unit cell is shown. (However, a 3 by 3 by 3 block of unit cells can be generated with the Multiscale Models tool.)
The checkbox settings control which matrix information is used to determine the number of copies and their transformations:
Many problems are due to information that is missing from (or incorrect within) the input PDB file. One way to validate symmetry information is to identify steric clashes with the Crystal Contacts tool.
Unit Cell does not generate multimers defined by BIOMT records, but this can be done with Multiscale Models or the command sym.
Unit Cell generates only a single unit cell; however, Multiscale Models can generate a 3 by 3 by 3 block of unit cells.