

 home
overview
research
resources
outreach & training
outreach & training
visitors center
visitors center
search
search
home
overview
research
resources
outreach & training
outreach & training
visitors center
visitors center
search
search


 home
overview
research
resources
outreach & training
outreach & training
visitors center
visitors center
search
search
home
overview
research
resources
outreach & training
outreach & training
visitors center
visitors center
search
search

Catherine L. Lawson1, Tom Goddard2, Matt Dougherty3, Hirofumi Suzuki4 Christoph Best5, Richard Newman5, Kim Henrick5,
1
RCSB - PDB
Rutgers University
2
Resource for Biocomputing, Visualization, and Informatics
University of California, San Francisco
3
National Center for Macromolecular Imaging
Baylor College of Medicine
5
EMDB - PDBe
European Bioinformatics Institute
The Electron Microscopy Data Bank (EMDB), established in 2002 at the PDBe, European Bioinformatics Institute (EBI), is a public repository for electron microscopy density maps of macromolecular complexes and subcellular structures. It covers a variety of techniques, including single-particle analysis, electron tomography, and electron (2D) crystallography. The EMDB contains experimentally determined three-dimensional maps and associated experimental data and files.
Chimera is the primary package used for visualization of EMDB data. Ongoing improvements to EMDB and Chimera are enabling maximum use of deposited map and atomic model data by researchers. Some of these developments are described below.
Searching EMDB within Chimera
To simplify the process of viewing EMDB maps and fit PDB models in Chimera EMDB is developing a search web service that is utilized by Chimera to directly search for maps by keyword, author, or accession number and allow direct download and display. This supplements the web browser interfaces to EMDB data.Chimera EMDB Search: Search results for keyword "microtubule". Search was made within Chimera and one of the resulting maps and fit PDB models was fetched.
Verifying Alignment of PDB and Electron Microscopy Databank Models
The Electron Microscopy Databank (EMDB) (Tagari, 2002) is designed to manage, organize and disseminate the structural data of macromolecules solved by 3-D electron microscopy, and is hosted at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/emsearch/index.html). Many of the volume maps deposited at the EMDB have structure counterparts in the PDB. Users can fetch corresponding data for visualization and analysis. At present, fitted coordinates entries in the PDB exist for 47 of the maps available in the EMDB. However, because of inconsistencies in transforming maps to the archived format used by the EMDB, the map and coordinates can be readily superimposed in only 21 of the 47 cases. Misalignments are identified visually using Chimera and its VolumeViewer extension. The figure below shows a well-aligned pair, where there are few clashes between structure and surface. The planned development of additional features within VolumeViewer, and perhaps even automated and qualitative detection of misalignment, would be welcome additions to this work. We look forward to our continued collaborations with the RBVI in this area.
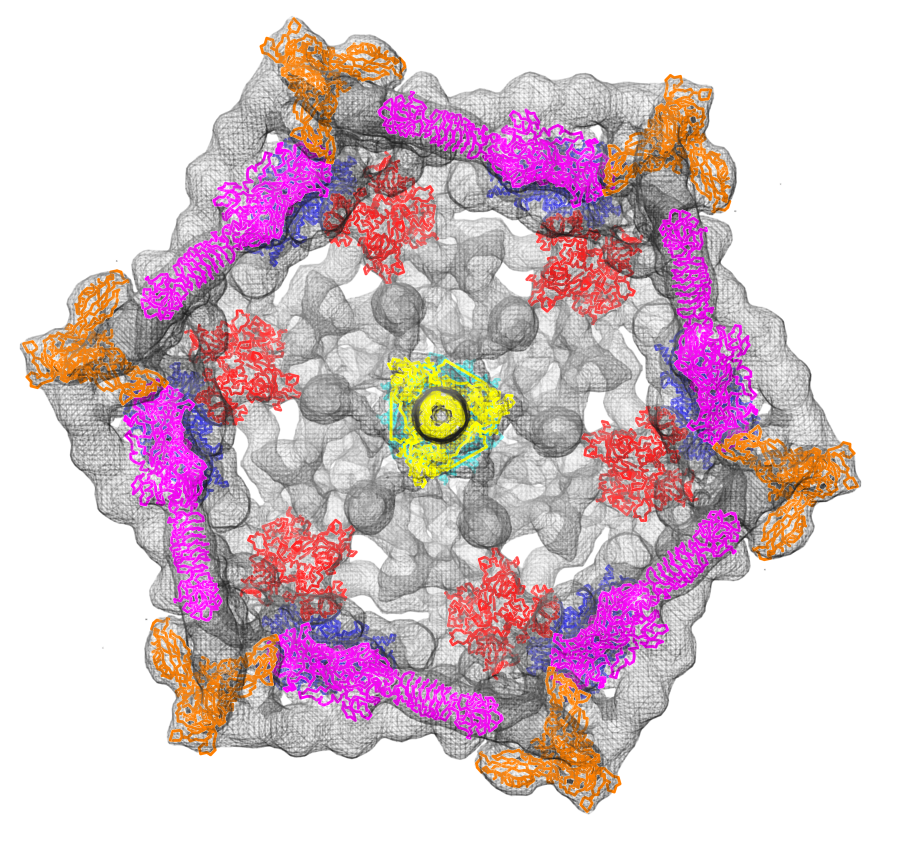
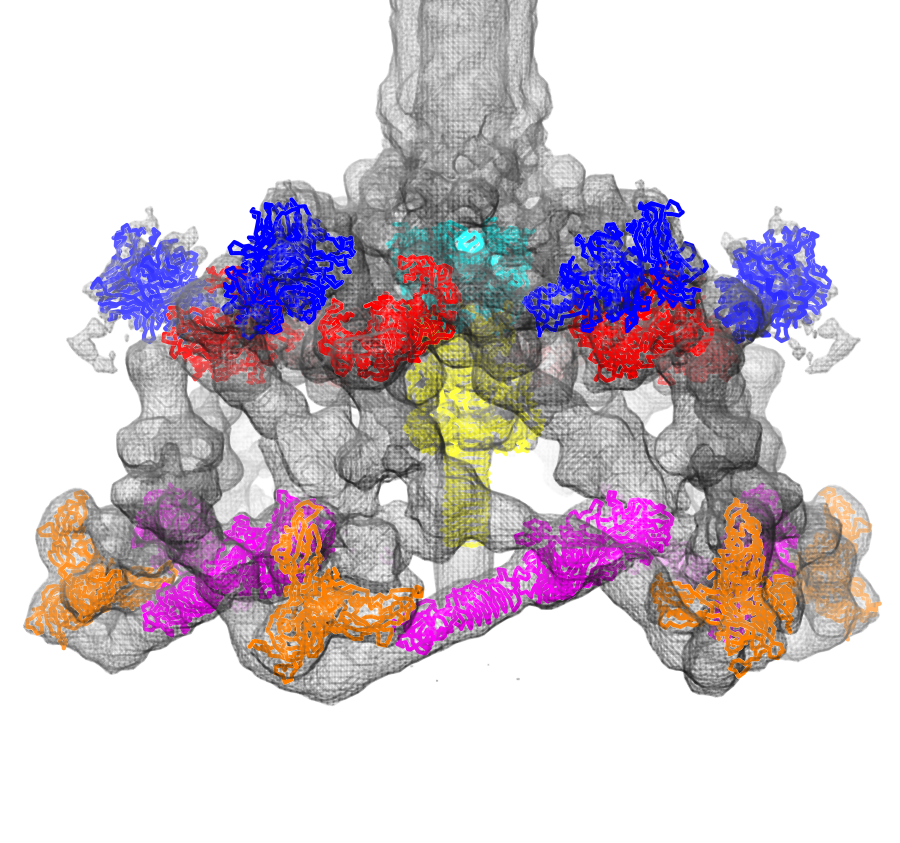
Fit atomic models: Structures of six proteins (PDB 1pdf, 1pdi, 1pdj, 1pdl, 1pdm, 1pdp) fit into a density map of the phage T4 baseplate (EMDB 1048).
Masked T-cell organelles: Color regions show masked objects in cell tomography data (EMDB 1155), lysosomes cyan, golgi red, centriole green, microtubules magenta, intercellular space yellow.
Mask Display and File Formats
Masks define objects observed in density maps. A mask is a 3-dimensional grid matching the size of the density map it corresponds to. Each grid point gives an integer index or bitmap indicating which object that grid point belongs to. While density map file formats can accomodate such data they lack provision for naming the objects, describing the index/bitmap convention, and assigning colors to the objects. The EMDB and Chimera developers are exploring options for mask file formats that encapsulate the desired data. Software for visualizing masks and using them to extract or analyze specific objects are also being developed within Chimera.Map Visualization on the Web
Visualizations on the EMDB web pages for each map are created with Chimera, and additional movies and Chimera sessions illustrating the data are provided by the EM Navigator web site. We are exploring use of advances in rendering quality such as per-pixel lighting, single-layer transparency, and silhouette edging for hand-created images that accompany each EMDB entry. Chimera scripts to automatically generate movies that slice maps, and session files that provide informative views have been developed and continue to evolve.Map Formats
We are designing and testing map file formats to overcome limitations in the currently used CCP4 format. Desired new capabilities include specifying symmetry, object names, colors and index format for masks, positioning matrices for fit atomic models, handling all commonly used data value types (unsigned 8-bit not supported by CCP4), including subsampled map copies for high-performance multi-resolution access to large maps. We are considering two alternatives: 1) adapting the existing MRC2000 format by appending XML meta-data describing symmetry, fits, and other parameters, or 2) using the high-performance extensible HDF5 file format. The first option maximizes compatibility with existing software while the second offer high-performance for large data sets.Thumbnail gallery of EMDB maps.
References:
- Tagari M, Newman R, Chagoyen M, Carazo JM, Henrick K. New electron microscopy database and deposition system. Trends Biochem Sci. 27(11):589, 2002. (PMID: 12417136)
Laboratory Overview | Research | Outreach & Training | Available Resources | Visitors Center | Search