

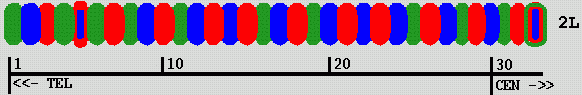
Mike fluorescently labels segments of chromosome 2L in a Drosophila cell nucleus using 3 fluorophors. This produces a color "bar code" pattern on the chromosome. Each labeled segment is roughly 80 Kbases of DNA. There are rougly 500 Kbases between adjacent labeled segments. He has tried up to 33 labeled segments covering most of the chromosome (about 20 Mbases). Using just 13 labeled segments has proved easier to analyse.
|
| Figure 1: Color bar code labeling Drosophila chromosome 2L |
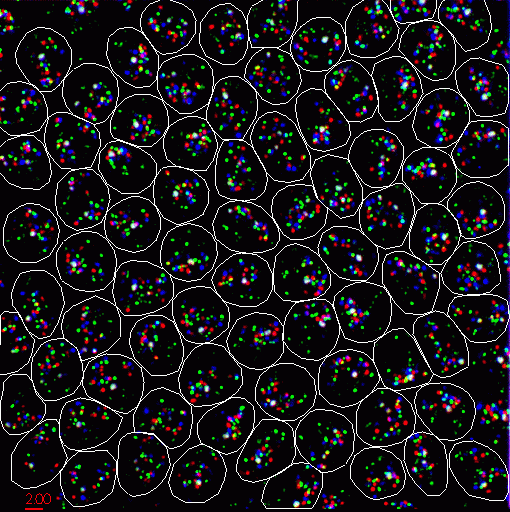
The labeled cells are viewed with a light microscope. 2D images are acquired at about 100 equally spaced depths. These images are stacked up to create a 3 dimensional data set (a couple hundred Mbytes). There are roughly 100 cell nuclei in one data set.
|
| Figure 2: Projection of 3D microscope data showing Drosophila cells with FISH labeled chromosomes. |
The next step is to trace out the chromosomes in each nucleus using the expected sequence of colored spots. There are 2 homologous copies of each chromosome in the nucleus. So two sequences of colored spots must be deduced for each nucleus. A program Mike wrote scans the image data and produces a list of the xyz positions of all the bright spots. For the data set where 13 segments are labeled, the goal is to number the colored spots seen in the images 1, 2, 3, ..., 13, and 101, 102, 103, ..., 113 for the two homologous copies of the chromosome. These numbers get recorded in the text file along with the xyz coordinates of the corresponding spots. He has done this by visually inspecting the sequence of z-planes to identify the expected sequence of colors.

|
| Figure 3: A single plane through a cell nucleus (june14.Bar12.1.n39) with labeled chromosomes. Quicktime movie (700 Kb) flipping through stack of image planes. |
It would be easier to trace the chromosomes with a 3D view of the data.
| ||
| Figure 4: Stereo pair showing a constant intensity surface of microscope data. Quicktime movie (1.4 Mb) of rotating isosurface. |
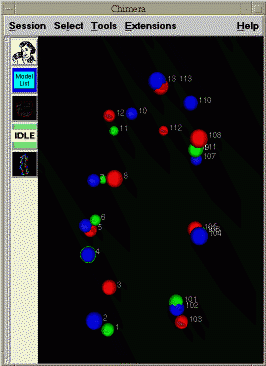
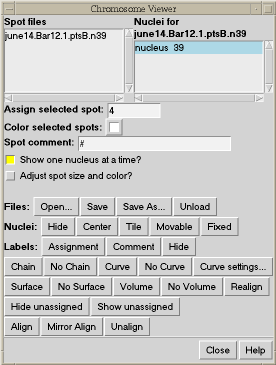
To facilitate assigning the chromosome segment numbers to the spots, spheres can be placed at the locations of bright spots. These are really atoms in Chimera and can be selected with the mouse. You can select one and type in a segment number. The segment numbers and spot xyz positions can be written to a file.
| 
|
| Figure 5: Spheres centered on bright spots in data with numbers indicating chromosome segment. | Figure 6: Chimera extension for tracing chromosome path. |
When the chromosomes have been traced, this Chimera extension can draw a smooth curve through them to better visualize the path of the chromosome.
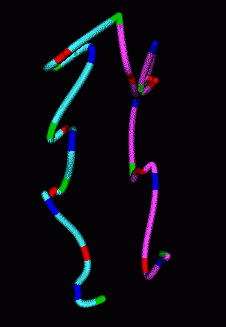
|
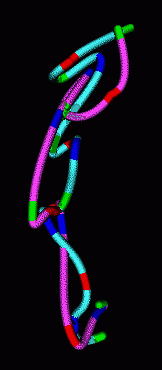
|
| Figure 7: Smooth curves representing paths of homologous chromosomes. | Figure 8: Minimum RMS alignment of homologous chromosomes. |
The homologous chromosomes often have similar looking paths. To assess how alike the paths are we can align the two models to minimize the root mean square distance between corresponding colored spots. The RMS is 1.12 microns in the example shown.
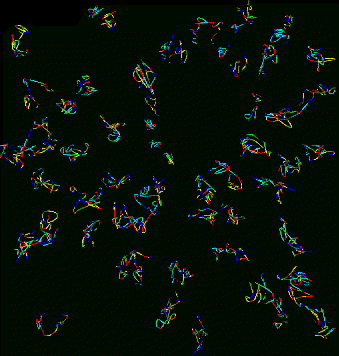
|
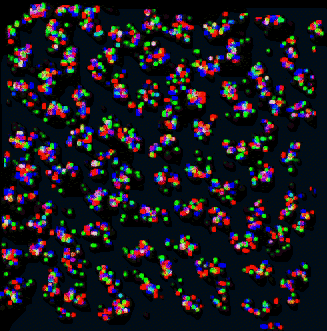
|
| Figure 9: Curves for 53 traced nuclei positioned as they occur in the sample. Not all nuclei in the data set were traced. QuickTime movie (2.2 Mb) rotating traced paths. | Figure 10: Projection of data corresponding to traced curves of Figure 9. The scale is slightly different then in Figure 9. QuickTime movie (3.1 Mb) rotating data. |
Does the spatial layout of the chromosomes vary from cell to cell in an organized way? The orientations may be correlated to the differentiated state of these embryonic Drosophila cells.
Above the microscope data is shown as a constant intensity surface. This 3D representation has advantages and disadvantages over flipping through the 2D stack of images. It shows the 3D geometry well, but you don't see as much detail in the shapes and intensities of the colored spots.
A 3D volume model in Chimera gives an improved view of spot shape and intensity.
| ||
| Figure 11: Stereo pair showing volume rendering of FISH labeled Drosophila nucleus. Quicktime movie (6.9 Mb) of rotating volume. |
The volume model is a projection of the 3D data set. The data is treated as partially transparent so points in back can be seen through the points in front. It is something like looking at a transparent glass model of the data.
It is useful to see where the chromosome lies relative to the nuclear envelope. Specific regions of the chromosome may be attached to the nuclear envelope and this could effect gene expression.
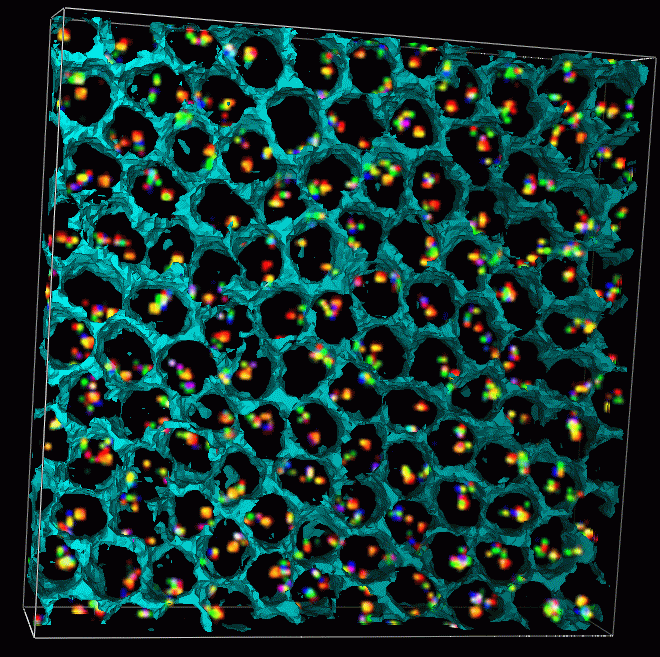
|
| Figure 12: Data set 02june01.2.3 that has been labeled with smaller 10 Kbase probes, consecutive probes separated by rougly 100 Kbases. The cells have been stained with DAPI that binds to all DNA. The DAPI signal is mostly contained inside the cell nuclei. The cyan surface shows an isosurface of DAPI at a low threshold and is a rough guide to where the nuclear envelopes lie. The nucleus occupies most of the volume of each cell at this stage of development. |
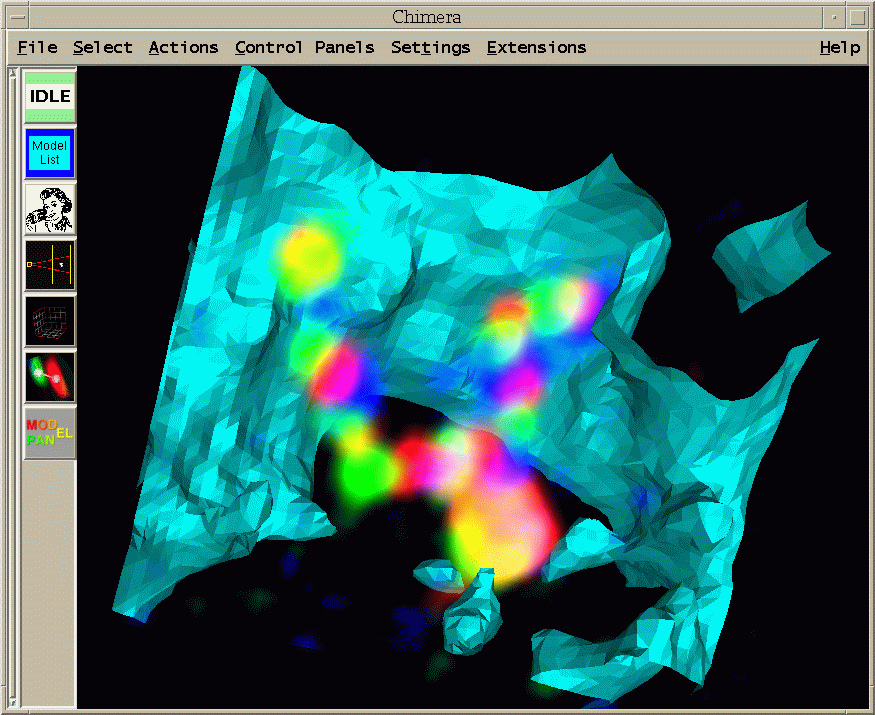
|
| Figure 13: Region showing single cell of 02jun01.2.3. Two sister chromosomes are seen. The large bright blob at the bottom is the labeled histone segment that contains many copies of the same histone gene. (Histone segment is probably much bigger than 10kb. Don't know how big.) The histone segment is labeled with all three fluorophors to provide a distinctive marker. I made a Quicktime movie of a different cell showing a range of perspectives. |
Laboratory Overview | Research | Outreach & Training | Available Resources | Visitors Center | Search