Tom Goddard
November 29, 2017
UCSF Biophysics 204A tutorial
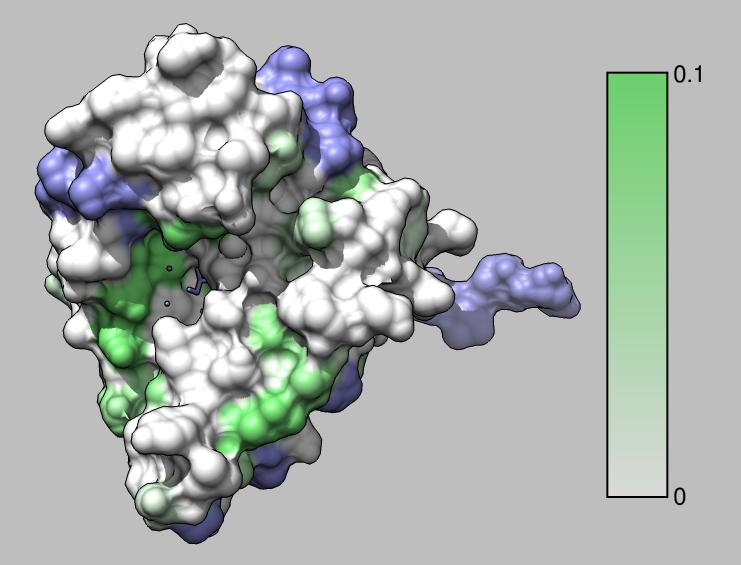
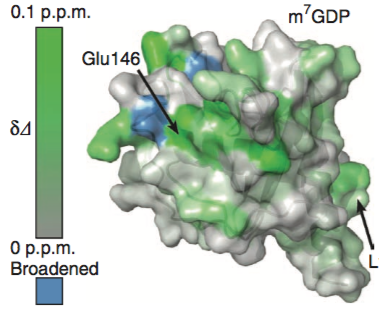
How to color the molecular surface of HSP90 by NMR chemical shift perturbation values using UCSF Chimera version 1.12.
First convert our chemical shift perturbation file that looks like
Assignment CSP
V201N-H 0.022
L56N-H 0.052
D142N-H 0.066
...
to a Chimera attribute file (documentation) that has residue number and chemical shift value and a minimal header like (note tabs are required):
#
# Chemical shift perturbations for HSP90, PDB 1ah8.
# Use this file to assign the attribute in Chimera with the Define Attribute tool.
#
attribute: csp
match mode: 1-to-1
recipient: residues
:201 0.022
:56 0.052
:142 0.066
...
The assignment in column 1 can be converted to a residue number with a Python script csp.py run in a terminal window, and an editor can be used to add the header lines saying it is a residue attribute.
python csp.py < shifts.txt > shifts_attr.txt

|
| Spin movie |
| Operation | Chimera menu entry |
|---|---|
| Open HSP90 protein, PDB 1ah8 | File / Fetch by ID. |
| Delete chain B | Select / Chain / B and Actions / Atoms / Delete. |
| Show surface | Actions / Surface / Show. |
| Read attribute file | Tools / Structure Analysis / Define Attribute. |
| Color using attribute | Tools / Structure Analysis / Render by Attribute. |
| Create color key on image | Render by Attribute panel button Create Corresponding Color Key |
| Save image | File / Save Image.... |
| Save movie spinning 360 degrees | Favorites / Command Line "movie record ; turn y 1 360 ; wait 360 ; movie encode ~/Desktop/1ah8_csp.mp4" |
| Save Chimera session | File / Save Session As... |
| Operation | Chimera menu entry |
|---|---|
| Gray background | Actions / Color / All Options |
| Shadows and silhouette edges | Tools / Viewing Controls / Effects |
| Reduce shininess | Tools / Viewing Controls / Shininess |
| Residue labels and arrows | Tools / Depiction / 2D Labels or use a photo editor program. |