Sequences can be viewed using either of the following approaches:
|
Standard residue names are converted to standard one-letter codes. In addition, certain nonstandard residues are assigned the same one-letter codes as closely related standard residues:
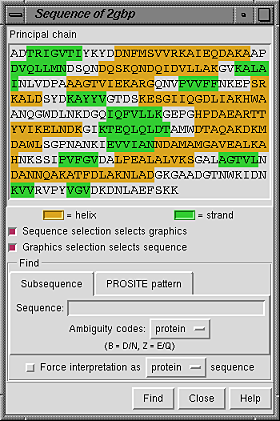
Placing the cursor over a residue in the sequence shows the residue number and name above the sequence. Stretches of color behind the sequence indicate secondary structure: goldenrod for alpha-helix and lime green for beta-strand (see the named colors). Protein helix and strand assignments are taken from the input structure file or generated with ksdssp.
When the checkbox designated Sequence selection selects graphics is activated, selecting a sequence segment within the sequence panel causes selection of the corresponding residues in the main graphics window. When the checkbox designated Graphics selection selects sequence is activated, a selection in the graphics window causes selection of the corresponding segment(s) within the sequence panel. Both checkboxes are on by default.
It is possible to find occurrences of a particular sequence string or pattern within the model; clicking the Find button at the bottom of the panel initiates the search.
Close dismisses the Sequence Panel; Help brings up a browser window containing this manual page.