Trying to summarize some of the regular tasks, with workflow details for the less obvious ones, in case anybody else is going to pick them up when I'm gone.
Whoever is doing the task may want to use different methods, which is fine! Undoubtedly my methods for some of these things are dumb and inefficient, and would be different if designed from scratch. But I wanted to provide enough detail so that it would be possible to carry on with the task in some manner.
Testing/Documentation← ChimeraX testing/documentation – I don't think I need to elaborate on this one, as you are all aware of it, know how to add/modify files in our github, and for guidelines on the general structure and format, you can view the existing files [User Guide]
Helpdesk
Citation tracking
Website
Licensing
← Chimera testing/documentation – not so much of this anymore, but similar to the above except that we use svn rather than git [User's Guide]
← ChimeraX and Chimera helpdesk (user email lists) – don't need to elaborate on this one either
← Citation tracking (both ChimeraX and Chimera) – I use Web of Science. Requires being on UCSF network directly or VPN.
- WoS queries (bookmarked URLs for my convenience, you could easily instead generate new queries with title containing program name and author ferrin or pettersen, or whatever else works): – these give ChimeraX and Chimera papers as well as their numbers of citations. For each paper, I click the citations number, sort by "recently added," and consult my personal notes on what these totals were the previous time I did citations, of to get an estimate of how far down each list I need to look. It's not exact, counts fluctuate for various reasons.
- For each citation: I search Pubmed in a separate Chrome window (better for cut-n-paste than Firefox, and link color indicates ones I've already visited), copy entry into relevant citations HTML file (see more details below), including a link to the paper. If fulltext, I search the paper for "chimera" and add brief comments on how Chimera(X) was used, and whether competitor programs such as Pymol, VMD, DSV were mentioned. If not in Pubmed, I copy the entry text from WoS and then use Google to find a link for the paper.
- The lists are in HTML files by program and year of publication, usually
based on the date first listed in Pubmed (sometimes I manually edit to print date)
in plato:/usr/local/projects/chimera/www/data/secure/chimera-citations,
names like chipap2024.html, chipap2025.html,
chix2024.html, chix2025.html. The
Chimera intranet index
has links for viewing these files as rendered in a browser.
Within the files, the text of each entry looks something like this:
The "font color" stuff is a relic from when I used to color-code entries differently, but now I still have it on each entry since it's used in my citation-counting scripts. The link text is just "link" or "open access" or "abstract". Within each file I have separate sections for our own publications, those describing plugins/interfaces, and everything else (the vast majority); each section is in reverse chronological order. Gray areas:<p><font color="black"> Structural basis for the interaction between the bacterial cell division proteins FtsZ and ZapA. Fujita J, Kasai K, Hibino K, Kagoshima G, Kamimura N, Tobita S, Kato Y, Uehara R, Namba K, Uchihashi T, Matsumura H. Nat Commun. 2025 Jul 1;16(1):5985. doi: 10.1038/s41467-025-60940-w. PMID: 40593603 <a href="https://www.nature.com/articles/s41467-025-60940-w">open access<a> (Chimera fitting, figures along with ChimeraX and Pymol) </font>
- if both Chimera and ChimeraX are mentioned by name and they seem to realize they are two different things, I add to both lists regardless of whether either paper is cited. So hopefully I don't put the same thing in the same file twice, but it can be in both a Chimera file and a ChimeraX file.
- if only one of the programs is mentioned by name but only the other one's paper is cited, I only add it to the list for the one whose paper was cited (e.g., if they say "ChimeraX" but only cite the Chimera paper, it goes in the Chimera list).
- if it's clearly wrong, like the paper is cited for protein blots, or they say "we used Pymol" with a citation to our paper and the figures look like Pymol, I won't count it, although it may get put into the file with HTML commenting out around it instead of the "font color" stuff so that I don't bother with it again.
- I note down potential featured citations for the respective homepages: high-profile (Nature, Science, Cell...) or otherwise interesting (description of plugin or protocol, great figures, or cool project), recent enough to show up on the homepage list, which is automatically assembled to show just the five most recent dates. More on this below.
- Every week, I also scan the latest issues of Nature and Science as soon as they come out, so that I can feature any citations therein right away; I add the day date to the Pubmed entry if it only has the month, so that they aren't bunched up on the first of each month (the featured citations are automatically sorted by date on the homepage).
- Citation-counting scripts are in the same dir as the citation files
(plato:/usr/local/projects/chimera/www/data/secure/chimera-citations):
- count – count all Chimera citations
- ncount – recent years of ChimeraX citations by year
- countx – ChimeraX citations by year.
- The graph that appears in the ChimeraX downloads & citations page is a screenshot of that Numbers graph, the file plato.cgl.ucsf.edu:/usr/local/projects/chimerax/www/preview/htdocs/images/citationsPerCalendarYear.png (updated weekly, SVN-committed)
- I note down potentially interesting papers for group meeting presentations
- many files for the homepage and immediate offshoots are in plato:/usr/local/projects/chimerax/www/preview/htdocs (ChimeraX) and plato:/usr/local/projects/chimera/www/preview/htdocs (Chimera)
- homepage and certain subsidiary pages are assembled nightly from templates and individual entries in the sections mentioned below. The template files include the quick links and the layout of other sections, and need to be SVN-committed after modifications: citationsTemplate.html, featuresTemplate.html, galleryTemplate.html, indexTemplate.html (homepage), newsTemplate.html
- for ChimeraX, the individual entries in these sections
(featured citations, news items, upcoming events, feature highlights, example
images) are individual HTML files named by date
(e.g., 07-05-2025.html)
in subdirs of the htdocs dir mentioned above:
citations, feature_highlights, gallery_samples,
news_items, events. These subdirs are actually logical links
to some other location and there is no versioning, the files within are used
automatically when the website is assembled.
Look at these files for examples of the formatting; with the exception of
feature highlights, they are all very simple, like just an HTML paragraph.
There is no mechanism to remove past "upcoming" events,
so their files have to be deleted manually.
Do not directly modify HTML files features.html, news.html, gallery.html, citations.html, as these are autogenerated nightly by assembling the individual entries. - for Chimera, it is much the same except the entire image gallery is under SVN in the ImageGallery subdirectory ...not that we care about the Chimera image gallery these days...
- To assemble preview websites (preview.rbvi.ucsf.edu/chimerax and
preview.rbvi.ucsf.edu/chimera), commands:
cd /usr/local/projects/chimerax/www/preview/htdocs; ./post_webpush
cd /usr/local/projects/chimera/www/preview/htdocs; ./post_webpush - I add new featured citations for both programs every week, news items primarily for candidate and production releases, upcoming events if we notice some ChimeraX workshop happening soon, feature highlights and example images as I have time and energy (not as often as I used to, invite others to chip in)
- ChimeraX Change Log for salient new features and updates – again, would welcome others chipping in, seems like it's mostly left to me and Eric, and I don't always remember to do it after updating the detailed documentation
- maintain/update miscellaneous other pages not in the User Guide:
- ChimeraX-related software – plugins/scripts not on Toolshed, web servers that provide files for ChimeraX viz/analysis
- ChimeraX vs. Chimera – ChimeraX missing features, reasons to not use Chimera
- ChimeraX tutorials index and some of the tutorials therein, although most are written by Tom G
- (various other ChimeraX pages have been maintained by Tom G: presentations, recipes, videos)
- maintain the Chimera(X) licensing information page
- that page says to use chimera@cgl.ucsf.edu for all commercial
licensing correspondence; in turn, it includes another address
(chimera@ucsf.edu) to provide the Pharm Chem team with a separate
mailbox that one or more responsible parties on their end
(currently Kimberly Cantero, others have been involved in the past)
can access, while keeping it separate from their other correspondence
Sept 2025: Greg converted chimera@cgl.ucsf.edu to a mailman list [archive] with me as the admin and me, Tom F, and chimera@ucsf.edu as members. When I'm gone, whoever's responsible for releasing license keys will need to replace me as admin and member. - bare minimum of what has to be done for the licensing process: wait for e-mail notice to release from Pharm Chem admin team (image below), generate license key using the link in the Chimera intranet, email license key along with instructions to licensee; more details about each of these steps in the example below
- other stuff I actually do: view all emails sent to chimera@cgl.ucsf.edu, update my personal Google spreadsheet for Chimera(X) licensing *...
- questions about software security and accessibility: I try to fend off the bureaucratic cover-my-butt drones who want official forms and company contacts regarding IT security and/or accessibility of the program. For use as a stock email answer, here is another Google doc (written up with the help of the team) about ChimeraX information security* In response to requests for filling out official forms (which can be ginormous and are usually mostly irrelevant), I basically say we are a small academic lab, not a software company, we're not going to do it, and they can simply decide not to license our software if that's not sufficient for their needs.
- questions about using the software: I try to answer these in the same way as questions on the mailing lists.
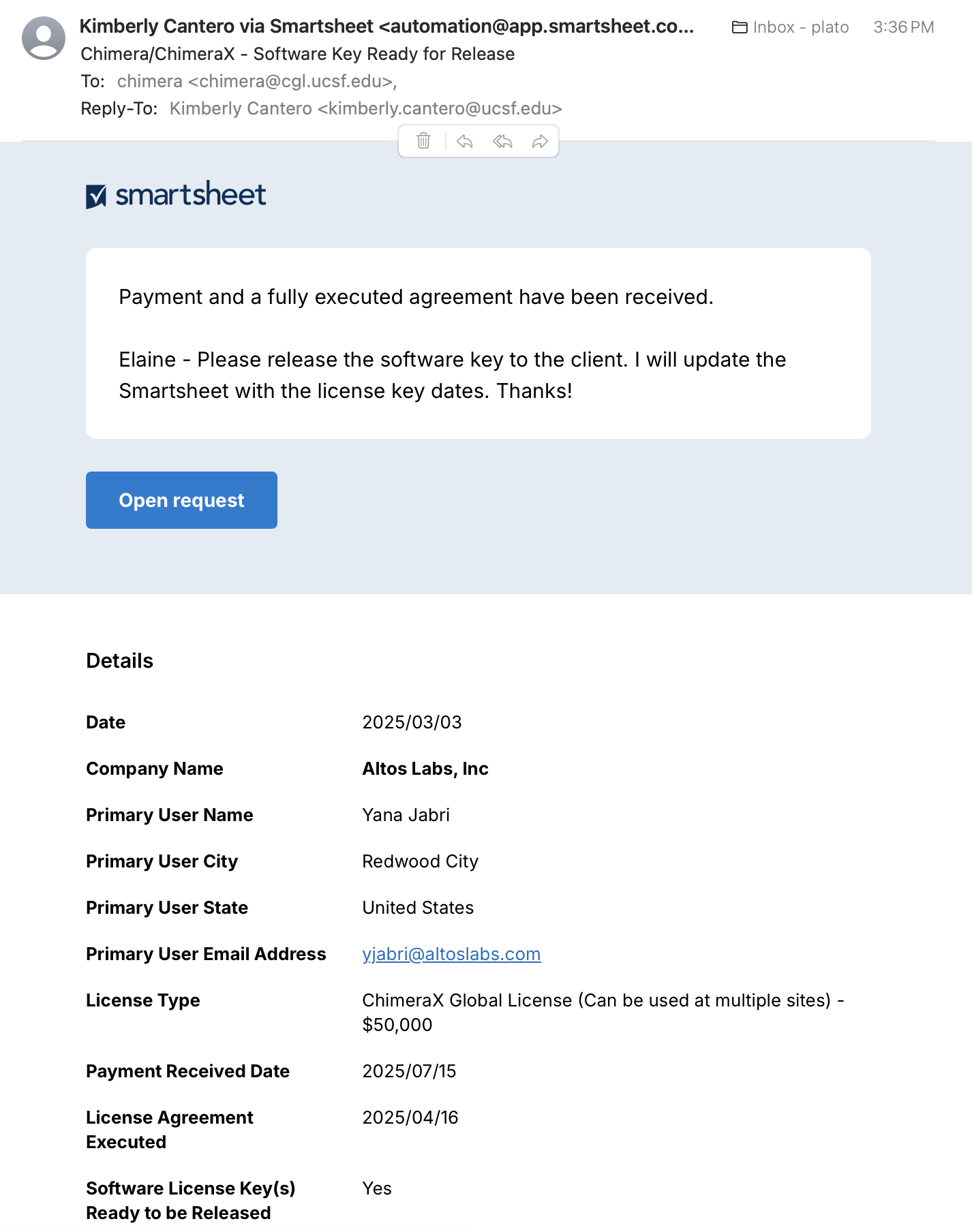
|
- received notification to release the license, email 7/15/25 (see figure). This notification format was developed with Kimberly Cantero in an iterative process and is meant to provide all the information needed to generate a license key. Usually it does, but not always.
- consulted
spreadsheet
*
to make sure it was in agreement with information in the notification.
Some stuff is wrong, e.g., state is not "United States,"
but I was going to put "global" as the site in the license key anyway.
There was already an entry for this license in the spreadsheet,
modified many times for all of the correspondence that had already taken place.
Examples of information added/modified in spreadsheet in response to emails: date of sending them an agreement and quote, date they return partially executed agreement, date OTMA returns fully executed agreement, date they provide a PO, any change in user name, location, and/or company name after they initially fill out the form to populate these parts of the agreement (since I need those to generate the license key), names/addresses of those who want to be CC'd on the license key email even though they're not the primary contact, etc.
There are also lots of “where's my license?!?” email messages even though they may not have returned a signed form or paid yet, or if they did pay, they did it wrong and we can't find it easily.
- in the Chimera intranet, there are links to generate a new license key and to view pre-existing license keys. I use the link to generate a new key, enter the information needed to generate the key (usually mostly in the notification email, but sometimes needs checking against the spreadsheet and/or old emails), and then save it to a text file
- I go to the relevant Google doc form letter * [ChimeraX] [Chimera] copy it, and paste into an email to the contact, CC myself, Tef, chimera@cgl.ucsf.edu, any others at the company that should also be CC'd according to my spreadsheet. Then I paste in the actual license key and edit number of users, or say it is global, and also attach the license key as a text file, then send it after doublechecking everything. In this case Tom L from Altos Labs had sent a message 6/27/25 requesting to be the primary user, but nobody on our end ever replied and Yana J had filled out the form with her own name, so that's what ended up in the license key. However, I CC'd Tom L on the email with the license key (sent 7/15/25).
- I change the color of the entry in the spreadsheet to light green after changing status and adding dates of release and expiration
- I delete most (but not all) of the past email correspondence for that license since my licensing mail folder is ridiculously huge.