ModelAngelo
Automated model building and protein identification in cryo-EM maps. Jamali K, Käll L, Zhang R, Brown A, Kimanius D, Scheres SHW. Nature. 2024 Nature. Apr 11;628(8007):450–457.
[github] (open-source MIT license)
- like our latest for ChimeraX, this is a Web of Science "hot paper"
 (published in past 2 years and citations from May/June 2025 put it in the
top 0.1% in the field of Biology & Biochemistry) as well as "highly cited"
(published in past 2 years and citations from May/June 2025 put it in the
top 0.1% in the field of Biology & Biochemistry) as well as "highly cited"
 (top 1% of field, not necessarily from the last 2 years)
– checked September 2025
(top 1% of field, not necessarily from the last 2 years)
– checked September 2025
- citations include those in high-profile papers from some of our expert users
(e.g., Veesler in Nature)
- machine-learning approach for automated building into cryo-EM density
- builds atomic models with similar accuracy to human experts
for proteins and for nucleotide backbones
- the latter may be especially notable considering that
AlphaFold3 performance on RNA lags that on proteins
(see commentary in Nature)
- outperforms human experts in identifying proteins of unknown sequence, i.e., it identifies the amino acid types from the density
[back to presentations list]
Background
- the number of new cryo-EM structures is growing exponentially
- most of the new structures in 2022 had resolutions better than 4 Å
- manual atomic model-building using 3D molecular graphics is typically time-consuming and requires expertise
- at resolutions worse than 4 Å it is challenging to avoid mistakes, and it is not uncommon for atomic models to contain errors
- if the amino acid sequence of the entity in the density map is not known, identifying it can be quite challenging, except at very high resolution; typically parts of the chain are built manually and then the sequence fragment is used in a database search to identify the protein
- the total number of atomic structures from high-resolution cryo-EM is relatively low (compared to the needs for training LLMs), at the time of this paper <13,000, many of them redundant
- automation is needed to remove bottlenecks and increase objectivity; previous work in this area includes DeepTracer and findMySequence
- "Although subsequent error checking and refinement remain necessary, ModelAngelo outperforms human experts in identifying unknown proteins and produces initial atomic models of comparable completeness to those obtained by human experts"
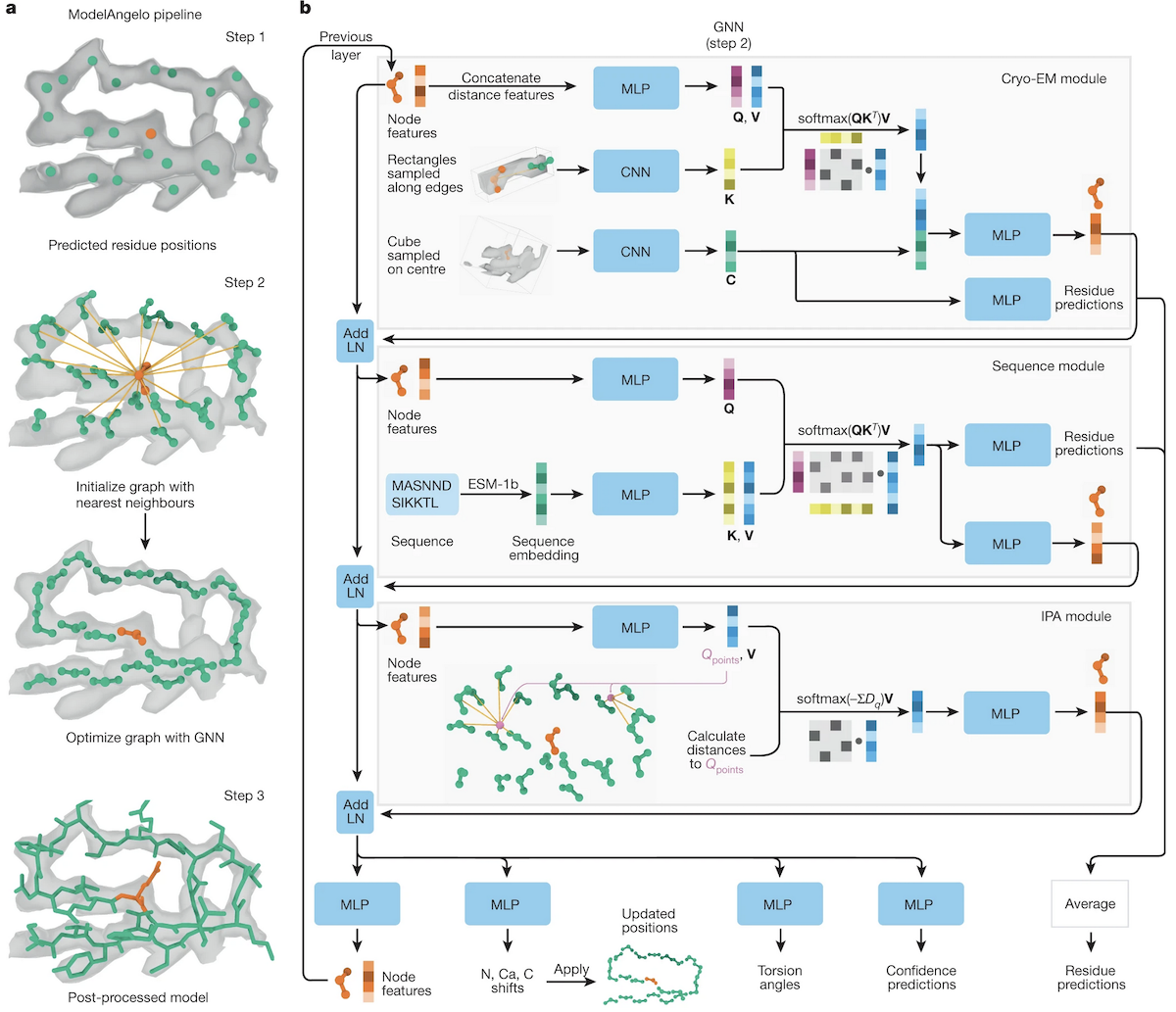
ModelAngelo Pipeline
- convolutional neural network (CNN) predicts whether each voxel contains Cα, P, or neither; edges drawn from each to 20 nearest neighbors
- graph neural network (GNN) involves passage through three modules,
each updating the residue feature vectors for the next step:
- cryo-EM module: features derived from the 20 rectangular boxes along the edges between a residue and its nearest neighbors, and a cubic box centered on the residue; multilayer perceptron (MLP) processing → residue type prediction
- sequence module (protein, not nucleic): user-provided sequences embedded with the language model ESM-1b (pretrained on many sequences, including homologs), more MLPs → residue type prediction
- invariant point attention (IPA) module inspired by that in AlphaFold2, four query points/residue → local topology, secondary structure
- postprocessing (more MLPs!) to generate a full atomic model, with 3 rounds of "recycling" graph optimization
Must have been delusional when I picked out this paper with 500 steps I don't understand.
Training
ModelAngelo was trained on 3715 map-model pairs: maps with resolution <4Å deposited before Apr 1 2022, resampled to pixel size 1Å, paired with PDB models covering the entire maps correctly. (Compare to 117 pairs for training findMySequence and 1400 for DeepTracer.)
ModelAngelo generates residue (amino acid) type predictions and converts them into HMM profiles for HMMER search against a set of sequences. The sequences are supplied by the user if the proteins in the cryo-EM experiment are known.
To enable model building for structures with unknown sequences, however, a version of ModelAngelo was also trained without the sequence module. That version also generates amino acid type probabilities and converts them into HMM profiles, but in that case, the HMMER search is against a larger proteome instead of just the user-supplied sequences.
Building Atomic Coordinates
- HMM profile-matched residues are assigned the corresponding residue types
- separate chains are connected on the basis of their assigned sequences and proximity
- chains shorter than four residues are removed
- a full atomic model is generated from the predicted positions and orientations of the residues and compared to predictions from idealized geometries. Backbone RMSDs are mapped to a confidence score between 0 and 1 corresponding to a linear range between 1.2 and 0.5 Å, respectively, and the score is written in the bfactor column.
Performance on Proteins
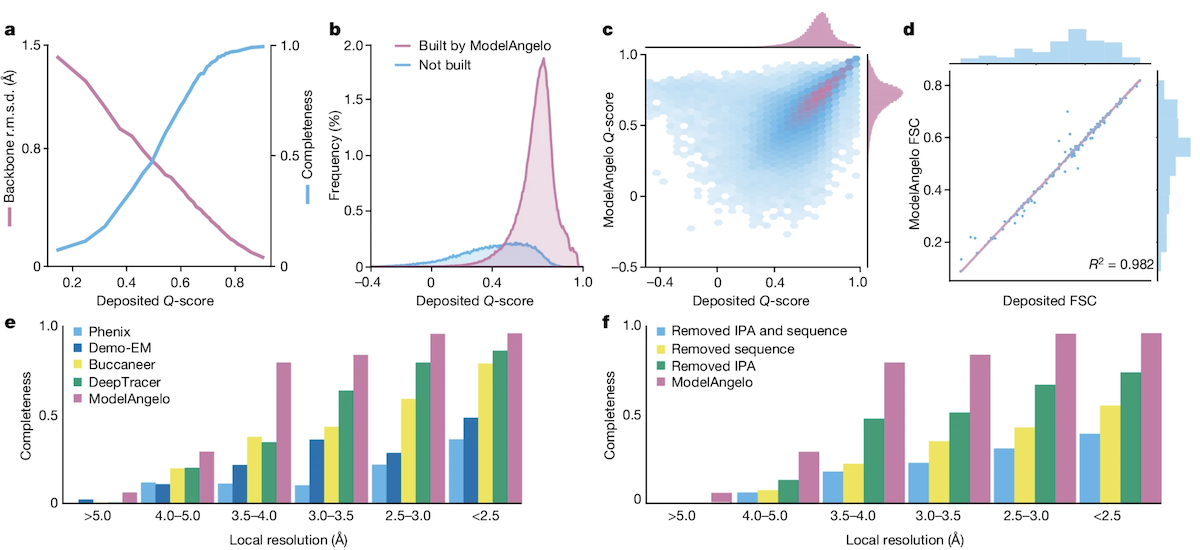
- tested on 177 structures determined to at least 4 Å resolution released between 4/1/22 and 2/9/23 with ≤ 30,000 protein residues, excluding icosahedral capsids (only a single asymmetric unit built), any with chains with >10% sequence ID to a training protein, or containing insertion codes or "other irregularities"
- runtimes per structure ranged 2-53 min
- output coordinates refined against the cryoEM map with a standard refinement cycle in Servalcat
- a-b: expected relationship between deposited Q-score (resolvability) and ModelAngelo completeness (fraction of residues with correct residue type and Cα within 3Å, blue) and RMSD (pink), with low RMSDs even at intermediate Q-scores and 77% of all test-set residues built
- c-d: deposited and ModelAngelo-built models are of about the same quality
- e: on 27 structures chosen for more detailed comparisons of automated model-building, ModelAngelo surpassed other methods for completeness and was able to build into lower-resolution areas (3-4 Å)
- f: all three modules contributed to ModelAngelo completeness
Performance on Nucleic Acids
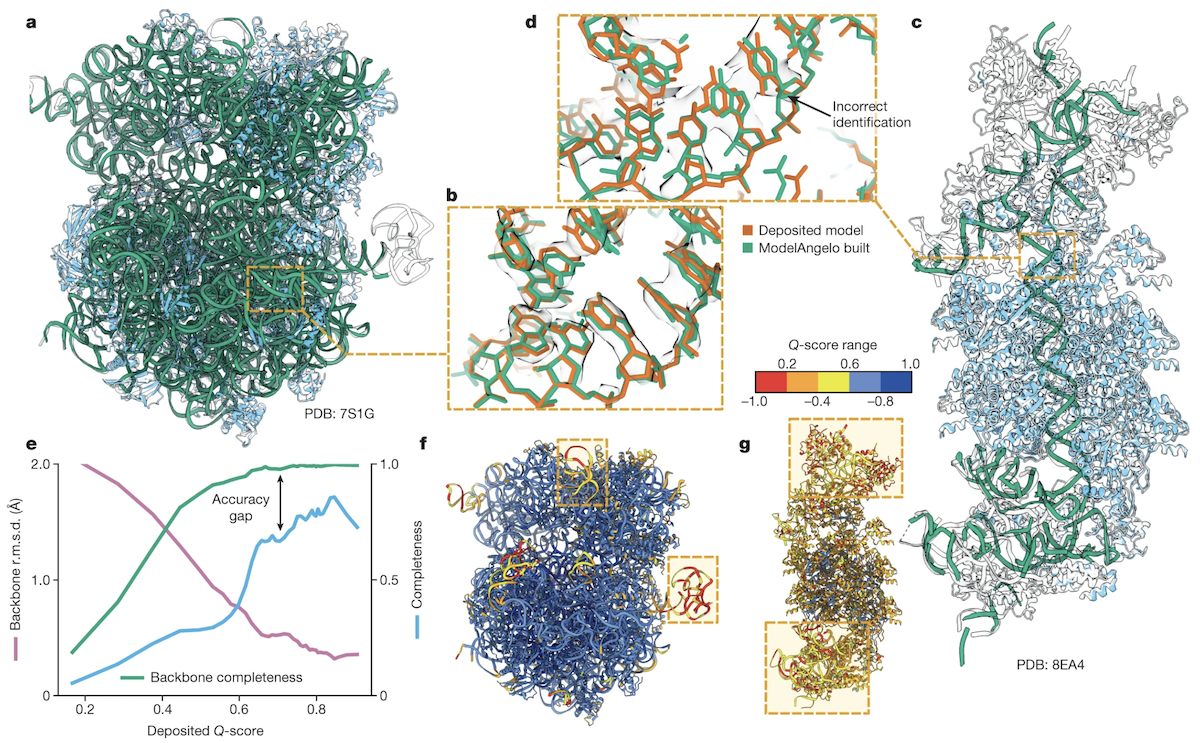
- only 103 NA chains in test set, but trends similar to proteins: ↓ RMSD and ↑ completeness for ↑ Q-score regions
- errors distinguishing sidechains A vs. G or C vs. T/U
- accuracy/completeness still better than DeepTracer and CryoREAD (subset: 3 ribosomes + CRISPR-associated transposome)
- a-b: E. coli ribosome, high-accuracy example (proteins blue)
- c-d: S. Hofmanni transposome, missing parts (black outlines) in low-Q regions, base assignment error (inset)
- e: RMSD (purple), backbone completeness (green), sequence completeness (blue) vs. Q-score
- f-g: deposited models for the same structures, boxes highlight low-Q regions
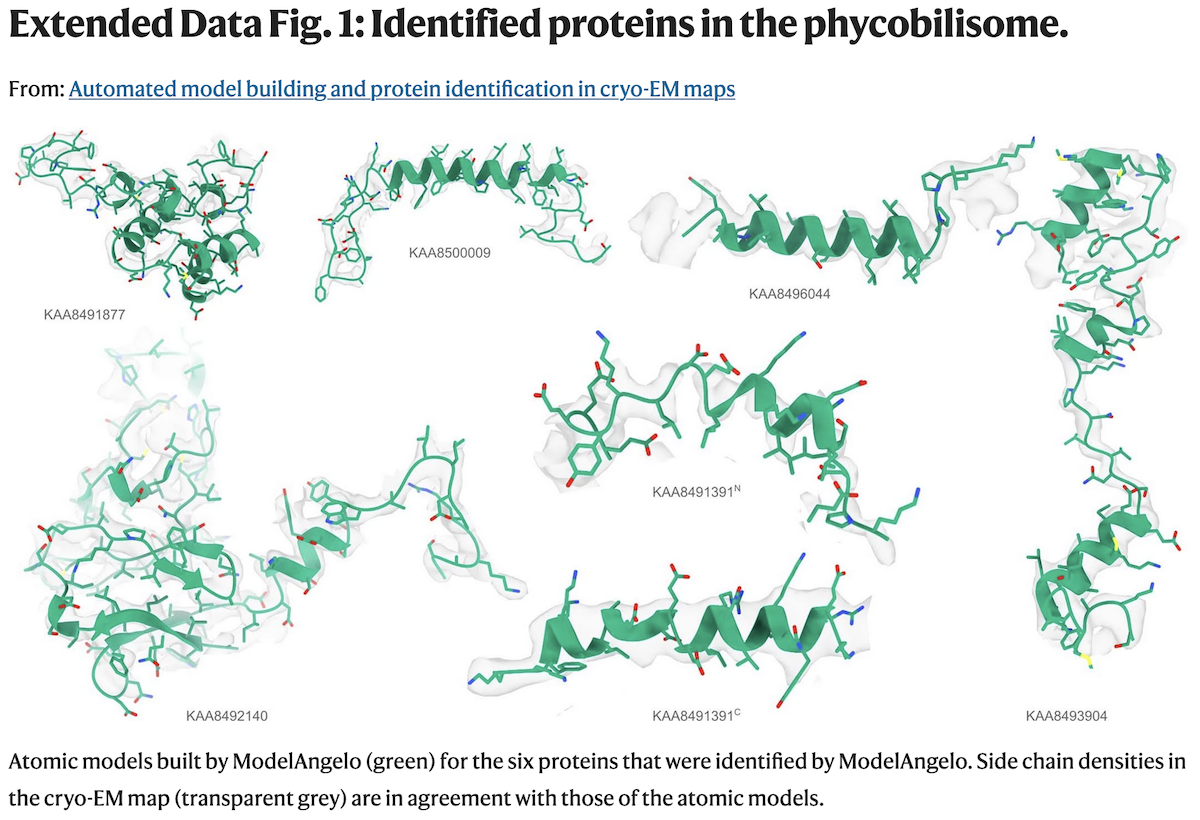
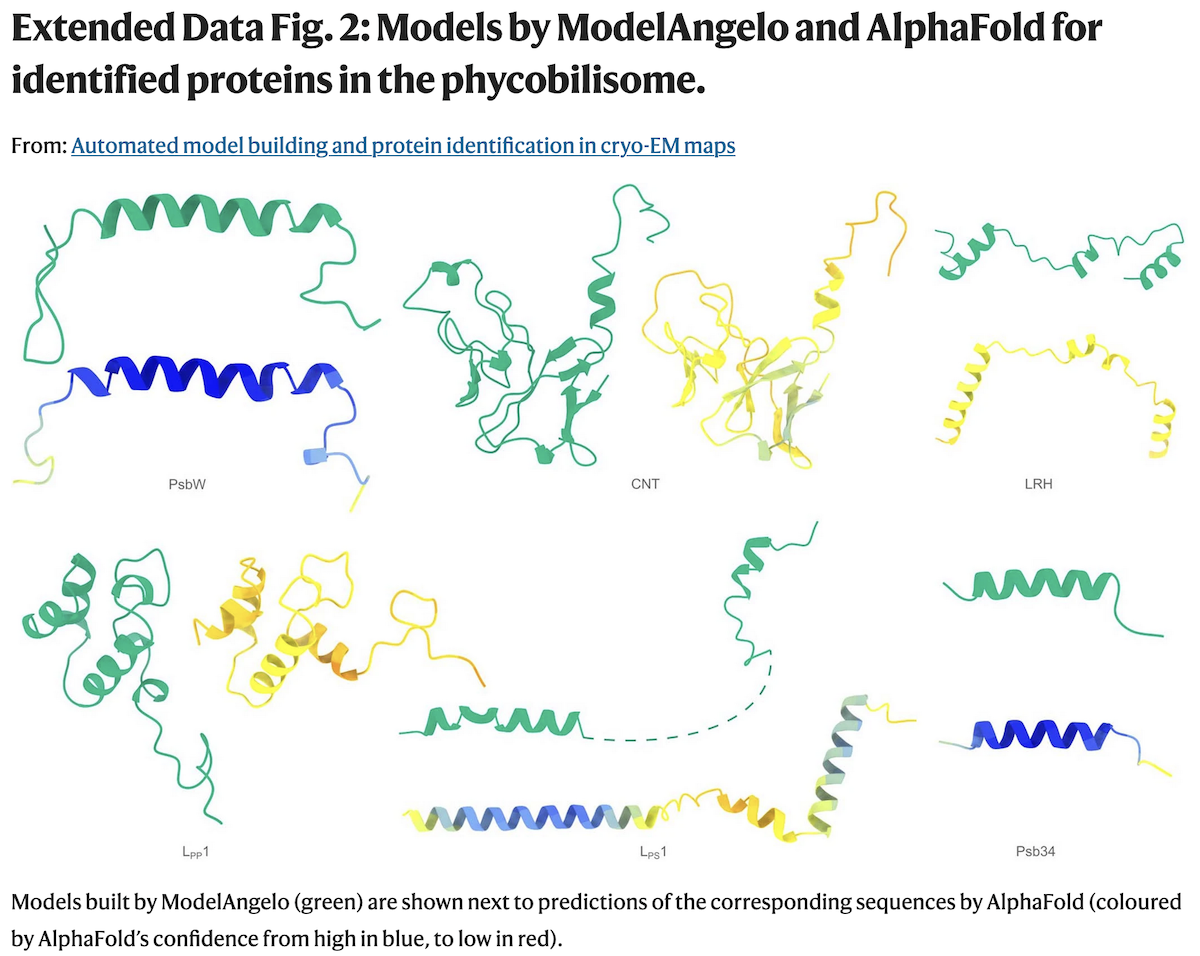
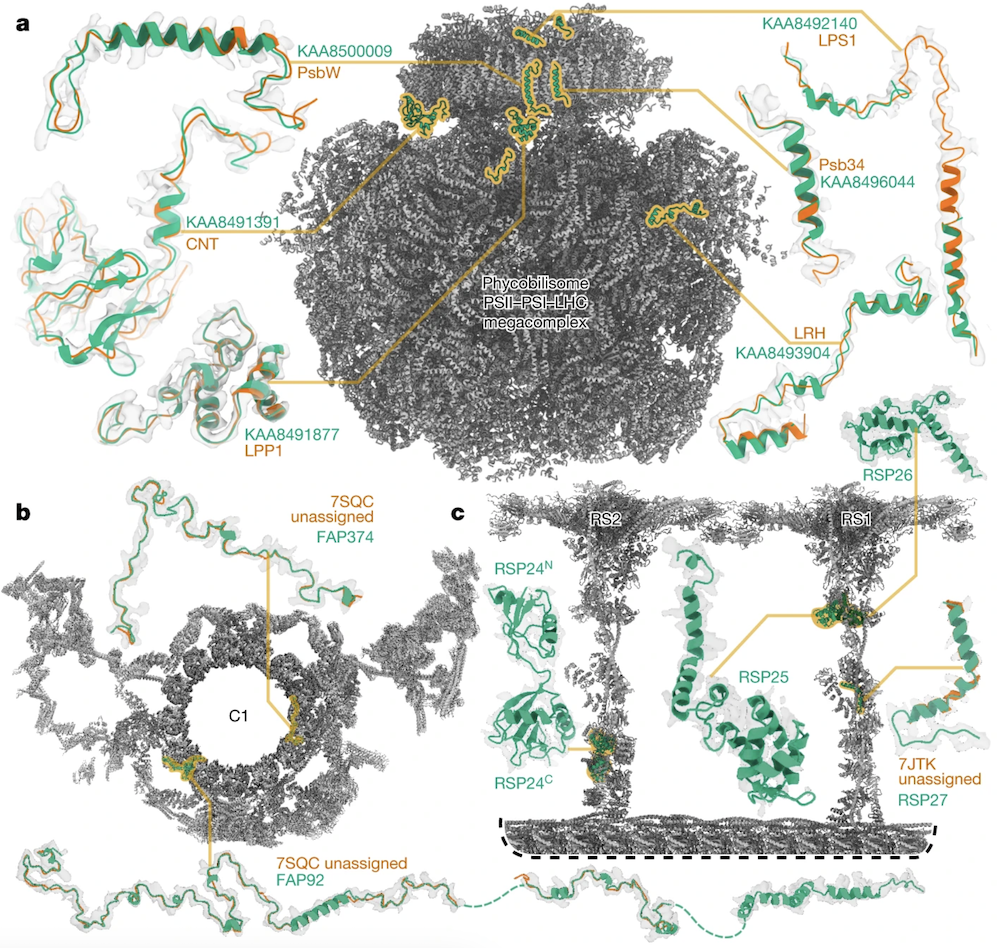
Examples of Protein Identification
- a: The phycobilisome-PSI-PSII-light-harvesting
supercomplex from red algae (158,730 residues in 81 unique protein chains).
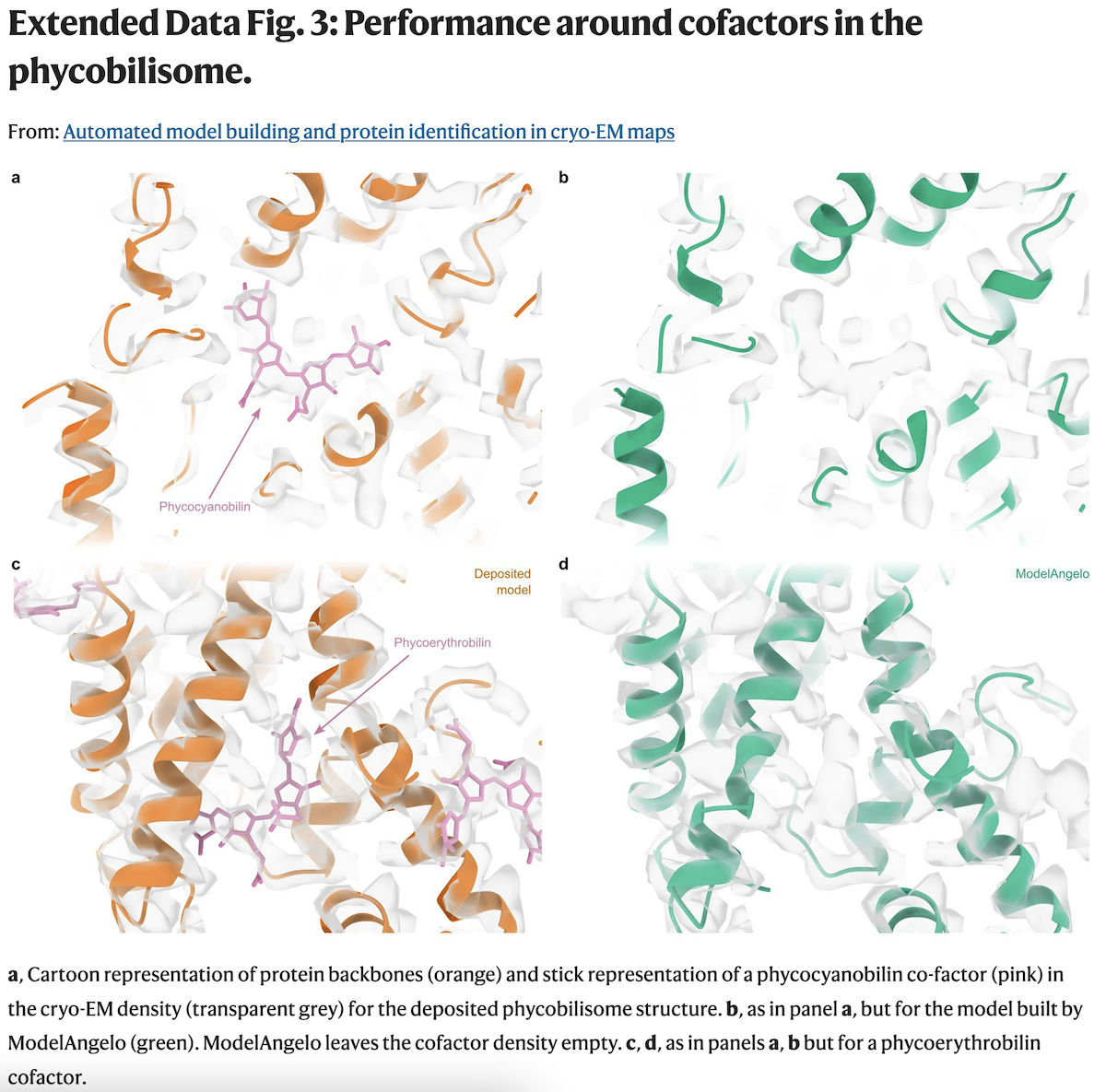
ModelAngelo did not try to build into cofactor locations, and w/o user
sequence input could identify 6 proteins (green) the original depositors could
not (orange), and do so consistently between pseudosymmetric copies;
findMySequence
identified 2 based on the deposited model, and a third
using the ModelAngelo backbone. After chain ID, all the sequences were input
and ModelAngelo run again (23 hrs) to get the best possible overall model
(gray, 110,742 residues), which had higher quality than the original deposition.
- b-c: C. reinhardtii ciliary axoneme, proteins built and identified by ModelAngelo (green) but not findMySequence. b: central apparatus microtubule C1, two chains deposited only as backbone. c: radial spokes 1 and 2 bound to a doublet microtubule, four proteins identified by ModelAngelo, only one of which had a deposited backbone