CombFold
CombFold: predicting structures of large protein assemblies using a combinatorial assembly algorithm and AlphaFold2. Shor B, Schneidman-Duhovny D. Nat Methods. 2024 Mar;21(3):477-487.
[github: code, Colab notebook, tutorial] [Code Ocean capsule]
- exhaustively enumerates assembly trees based on AlphaFold2-predicted dimers and (some) higher-order multimers ...they use "AlphaFold2" and "AlphaFold-Multimer" interchangeably...
- outperforms AlphaFold-Multimer (AFM) alone on benchmarks
- can incorporate distance restraints from crosslinking mass spectroscopy (XL-MS)
See also:
- AlphaLink: AlphaFold2 modified to incorporate experimental distance restraints
- MoLPC (Modeling of Large Protein Complexes): assemblies from AlphaFold2 dimers and trimers by Monte Carlo tree search [CoLab notebook] (mainly applicable to homomultimers, success rate ~30%)
[back to paper list]
Background
- Structures of large complexes are challenging to determine and/or predict:
- molecular machines exist in multiple states
- states can be fragile or otherwise difficult to prepare in needed quantities (for experiments)
- Existing prediction methods:
- Deep learning – AlphaFold-Multimer has a success rate of 40-70% for 2-9-chain complexes with up to 1536 residues, but for very large complexes, computational costs become prohibitively high, convergence to an accurate result becomes more difficult, and few solutions are provided after convergence
- Integrative modeling – relies on experimental data (XL-MS, FRET, coevolution, cryoEM, SAXS); applicable to very large and heterogeneous complexes, but associated methods require significant expertise
- Protein-protein docking – produces high numbers of pairwise solutions, but scoring functions are not that successful in identifying the correct ones (correct answer in top 10 ~25-40% of cases); if many pairwise solutions are retained, there is a combinatorial explosion of potential solutions for higher-order multimers
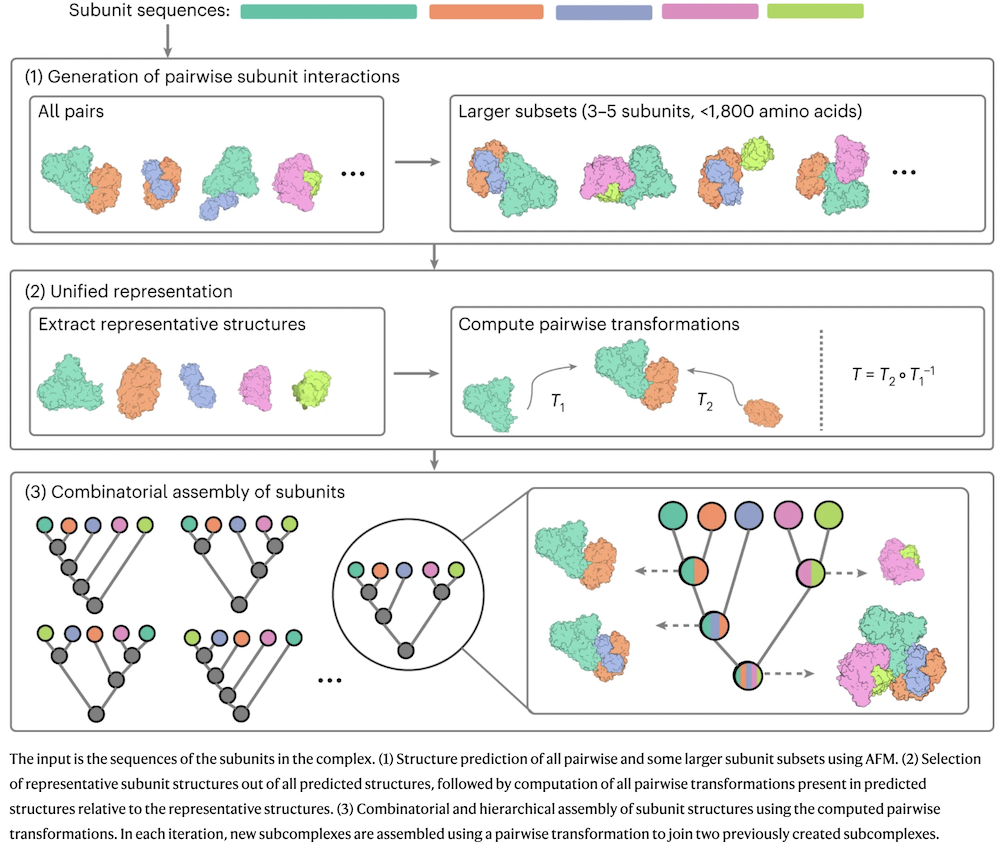
CombFold Overview
- run AFM on all pairs + three additional 3-5-mers for each subunit
based on that subunit's highest-confidence dimer interfaces
- a subunit can be a chain or chain segment; constraints enforce connectivity of consecutive segments (discard if >3Å x N)
- for each subunit, choose the structure with highest average pLDDT
as the representative
- calculate transformations (based on high-pLDDT residues) to generate each modeled interface from its representative structures
- each transformation (pairwise interaction) is associated with a score based on PAE (more later...)
- assemble the known or trial stoichiometry combinatorially, in each cycle discarding sets with worst clashes, constraint/restraint violations, or scores (slide ↓)
- confidence (overall score) is a weighted sum of the scores from the pairwise transformations used
Step 3 Filtering Example
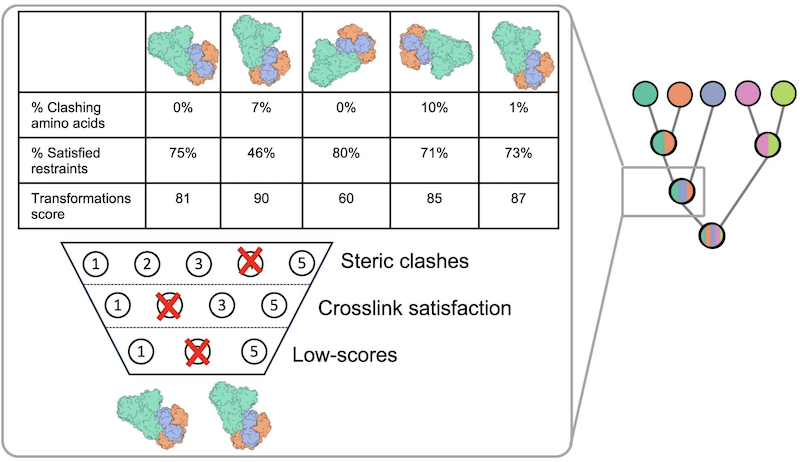
- to limit combinatorial growth at each stage of assembly, complexes containing the same subunits are Cα-RMSD-clustered and only the N best-scoring ones are kept (N=100 in the presented benchmarks)
- backbone atoms with pLDDT>80 evaluated for clashes, namely center penetrating >1Å into surface of another subunit; subcomplex removed if >5% of a subunit's backbone atoms clash
- overall score (confidence) is a weighted sum of the pairwise transformation scores
- XL-MS restraints are weighted based on experimental confidence (e.g. false discovery rate) and the average pLDDT of the crosslinked amino acids; restraint satisfaction ratio = Σ(satisfied)/Σ(all) is multiplied by the confidence score for filtering purposes
Performance
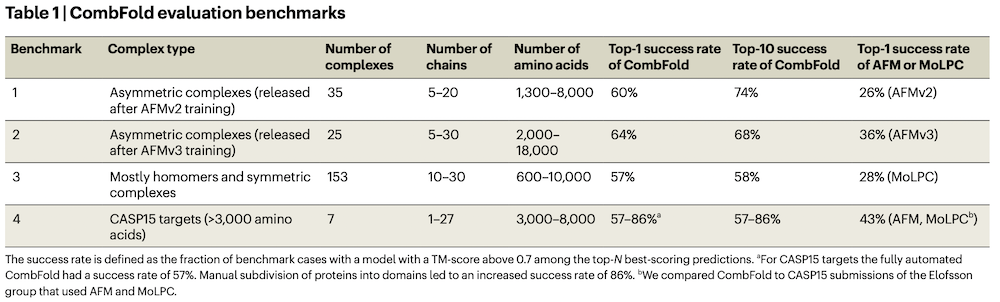
- Top-1 success 62%, Top-10 72% for Benchmarks 1 & 2 combined
- AlphaFold2 runs were performed with ColabFold, default parameters, no templates
- CombFold used AFMv2 for Benchmark 1, AFMv3 for Benchmark 2
Complexes in Benchmarks 1 & 2
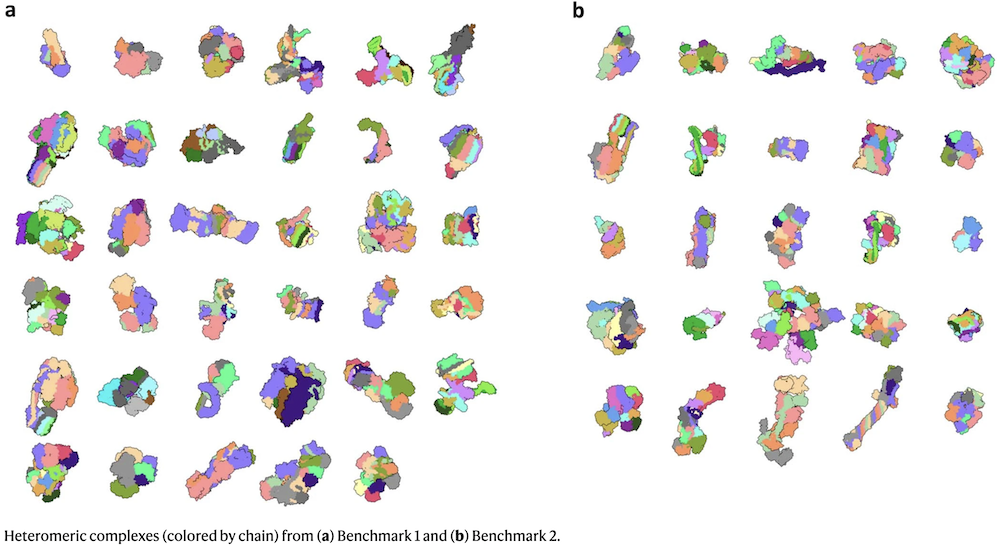
- Benchmarks 1 & 2 used entire chains as the subunits, except that long chains in five complexes of Benchmark 2 were split evenly into shorter segments until every pair could be predicted by AFMv3.
Benchmark 1 Details
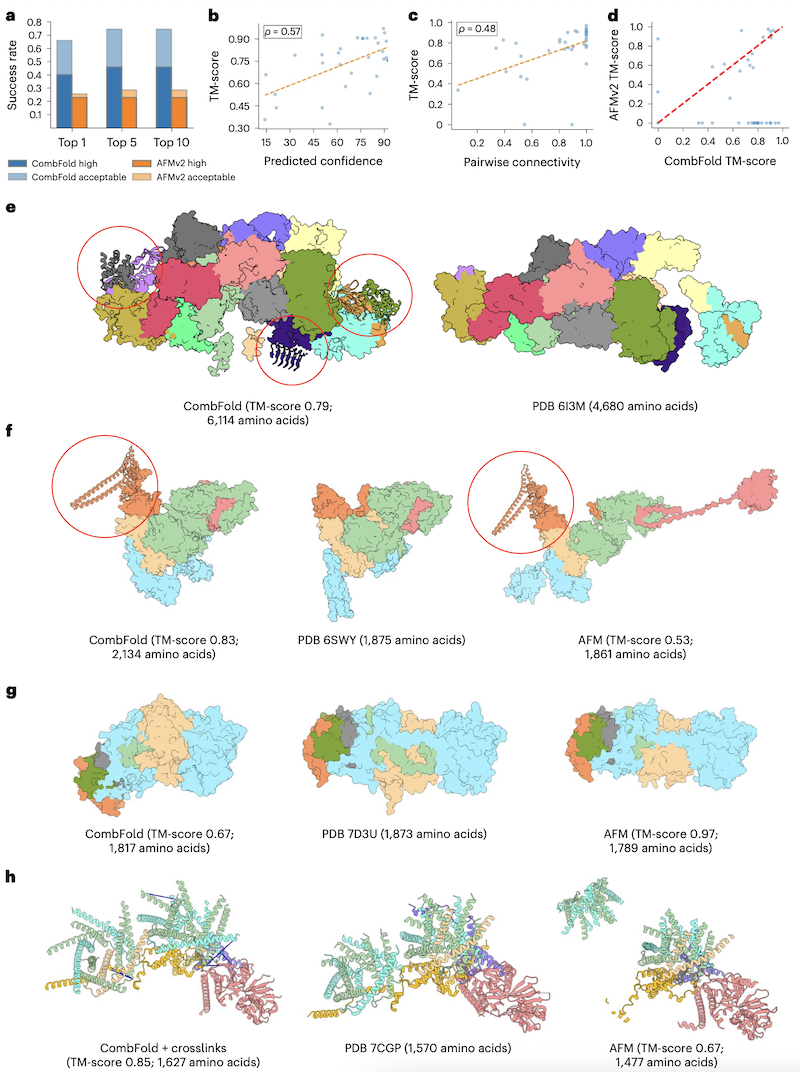
- a: CombFold (using AFMv2) outperforms AFMv2 alone; high indicates TM-score >0.8, acceptable TM-score >0.7–0.8
- b: TM-score correlates with predicted confidence
- c: TM-score correlates with correct chain-chain interfaces (pairwise connectivity is what fraction of the complex is in the largest connected component if nodes are subunits and accurately predicted interfaces are edges)
- d: CombFold (using AFMv2) outperforms AFMv2 alone
- e,f: CombFold gives more residues modeled (ribbons, red circles) than in the PDB, 20% more on average across Benchmark 1 counting those with pLDDT>0.5; the complex in e "could not be assembled directly" using AFM (7486 residues)
- g: example where AFM outperforms CombFold; the representative
structure chosen for the light-blue subunit has the wrong relative
orientation between domains
Restraints (not used for results in Table 1): - h: CombFold+restraints outperforms AFM, but without these 12 XL-MS restraints, the CombFold model was inaccurate too (TM-score 0.57). AFM does not allow specifying restraints, whereas AlphaLink does but is limited to <3000 residues.
Benchmark 1 Runtime Analysis
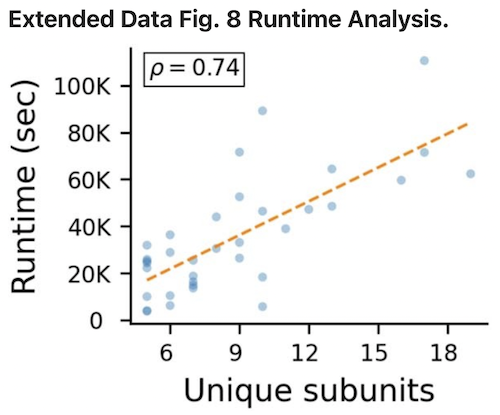
- 24 hours = 86,400 seconds
- NVIDIA A30 with 24GB memory
- ColabFold jobs were not run in parallel, but could be in principle
- runtimes are lower for homomeric complexes (e.g., Benchmark 3)
- assembly time is trivial (up to a few hundred seconds) compared to interface prediction
Benchmark 2 Details
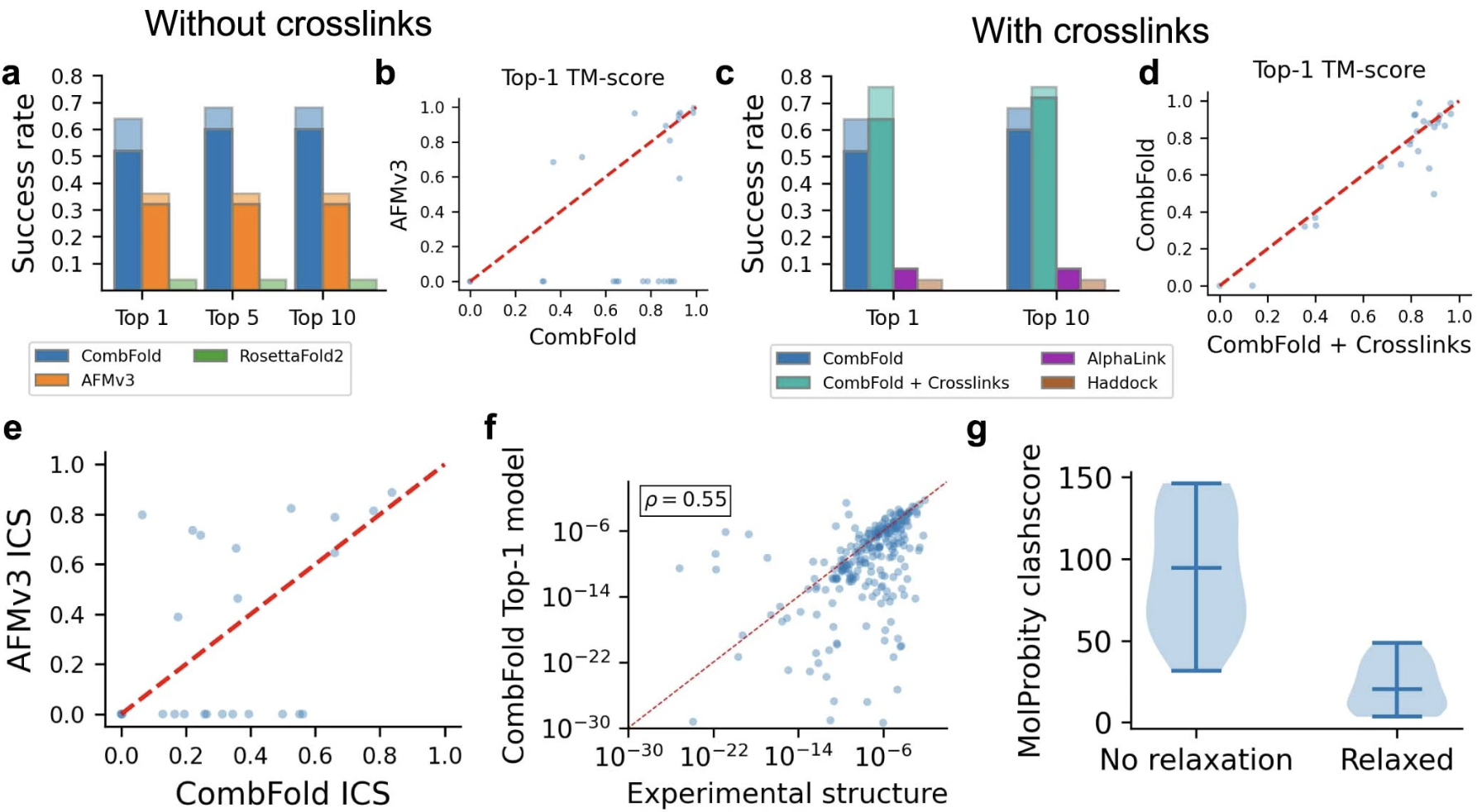
- a-b: CombFold (using AFMv3) outperforms AFMv3 alone; RosettaFold2 performed poorly, only 4/25 complexes assembled, only 1 with TM-score >0.7 in top 10 models
- c-d: with simulated crosslinks (slide ↓), CombFold 76% success compared to AlphaLink 8%, HADDOCK 4%, CombFold without crosslinks 64% (Table 1)
- e: TM-score represents global accuracy, whereas ICS (interface contact similarity) focuses on the interfaces; CombFold gives lower ICS scores than AFMv3 primarily because it uses single representative structures instead of those specific to each predicted interface. However, ...
- f: kD predictions with PRODIGY based on CombFold structures correlated well with those based on the experimental structures
- g: Molprobity clashscores could be significantly reduced by Amber minimization (plot shows sample of 17 structures) with little change in structure (Cα RMSD <1Å for all targets in Benchmark 2)
Benchmark 2 Details (cont.)
- Simulated crosslinks:
- From all pairs of lysine amino acids on different chains with a Cα-Cα distance <25Å, randomly sample 10%.
- To simulate noise, add 5% false detections of K-K pairs with
Cα-Cα distance >40Å.
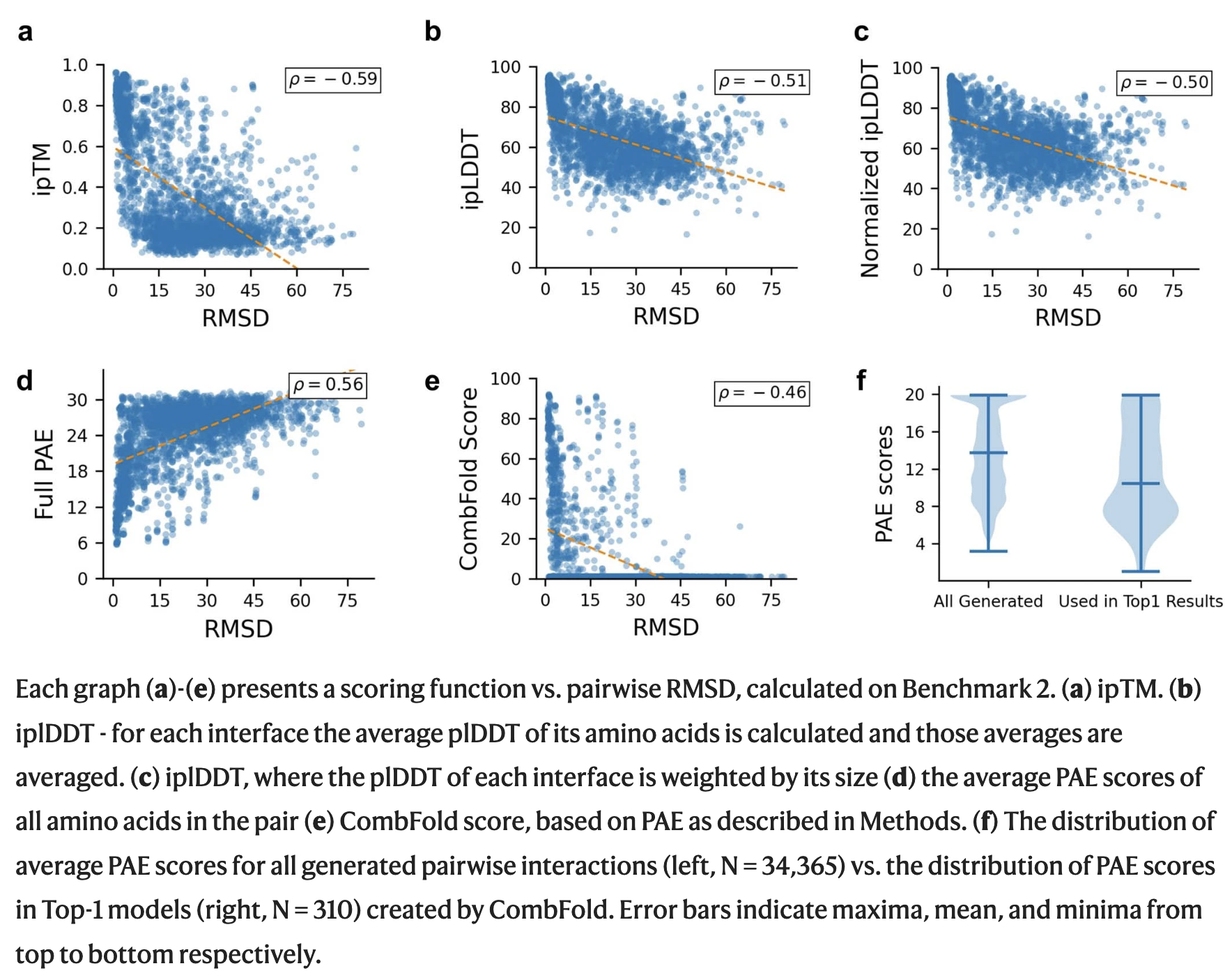
- Different pairwise scoring functions:
- AFM PAE score is calculated for every pair of residues in a structure, ranging from 0 (best) to 30 (worst)
- CombFold transformation score = max{1,100–P2/4} where P is the average PAE value of the two subunits
- this score ranges from 1 (worst) to 100 (best),
emphasizing small differences in the best (low-PAE) structures
- all scores correlated similarly with Cα RMSD, but (e) "the advantage of our PAE-based score is that incorrect interfaces consistently have low [worse] scores"; (f) those in the CombFold top-1 assembly have lower [better] PAE scores
Benchmark 3 Details
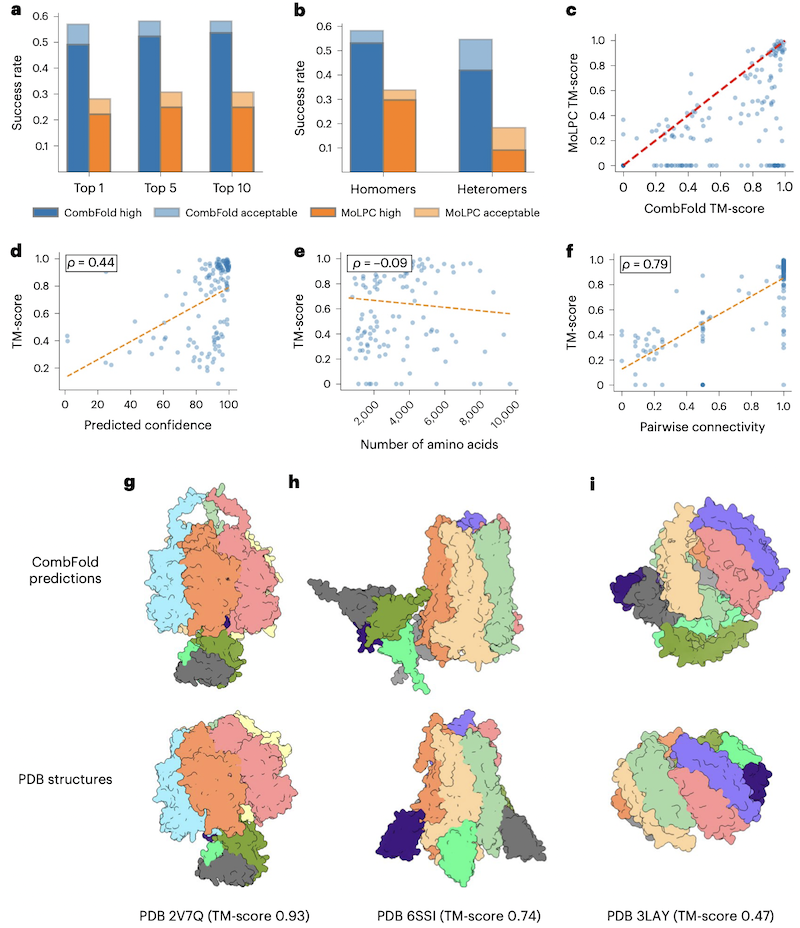
- a-c: CombFold outperforms MoLPC (top-1 57% vs. 28%, top-10 58% vs. 31%). The authors attribute this to CombFold's use of multiple AFM models, the highest-confidence prediction per subunit and emphasis on highest-confidence pairs, a more exhaustive combinatorial search, and effective winnowing by clustering and confidence scores that correlate well with accuracy.
- d,e: TM-score correlates with the predicted confidence but not with assembly size
- f: TM-score correlates with correct chain-chain interfaces. This correlation is higher than for Benchmark 1 (0.48) because in the assembly of homomeric structures, CombFold relies on fewer pairwise interactions; in other words, CombFold is limited by AFM's ability to predict the interfaces (reported success rate of ~60% in predicting pairwise interactions) and this limitation is more obvious in the case of homomers.
- g-i: examples with different TM-scores
Predictions for Complexes without Known Structure
- Complex Portal contains manually curated information on stable macromolecular complexes
- CombFold with AFMv3 was used on the 28 complexes with: >5000 residues, known stoichiometry, lack of homology to experimentally determined structures (≥ 66% of chains with ≥ 30% sequence identity)
- high-confidence models were generated for seven of these complexes
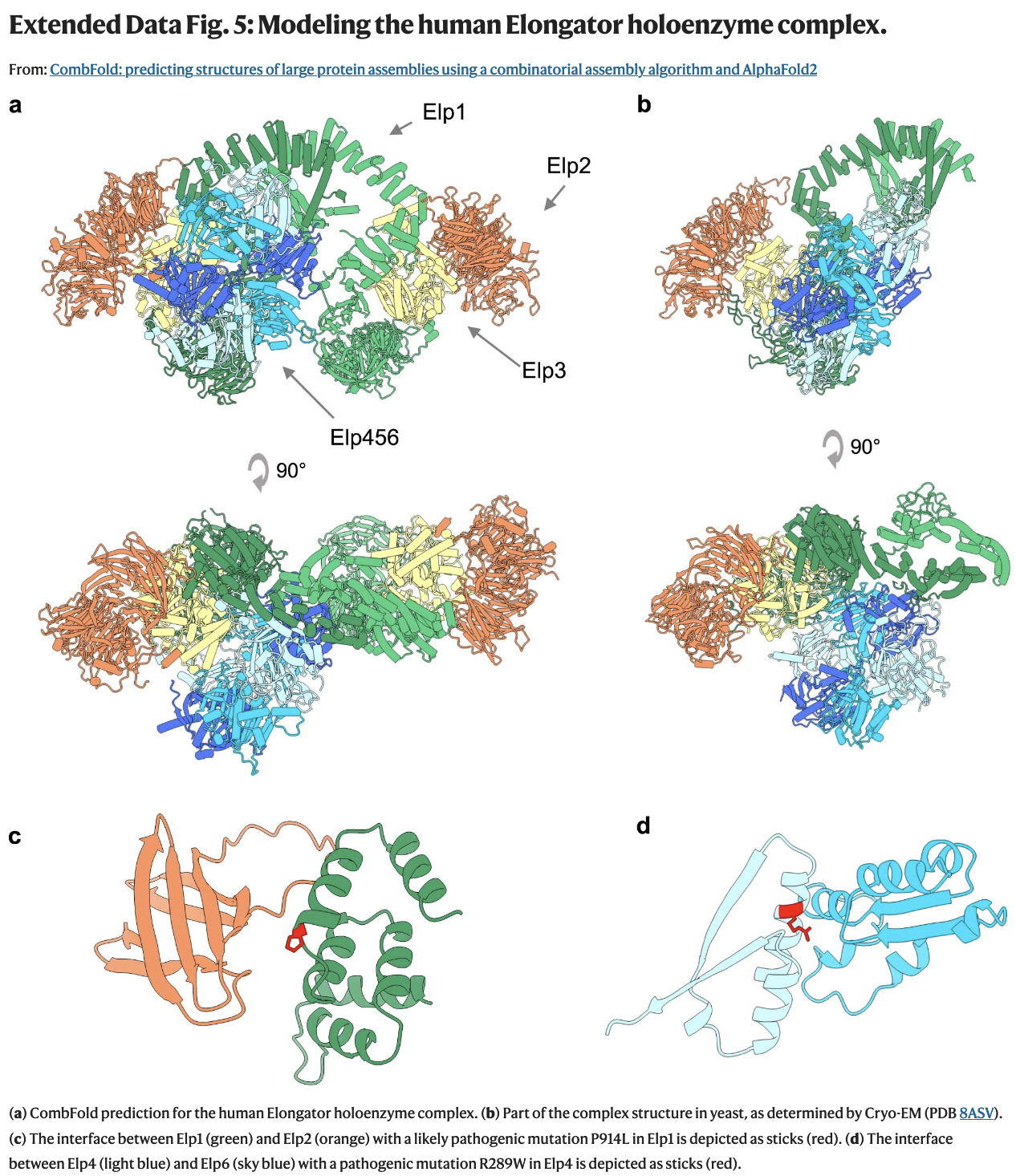
(Elongator + those on the next slide)
- human Elongator prediction (a) consistent with recently determined subcomplex from yeast (b)
- ten pathogenic or likely pathogenic missense mutations in Elongator proteins according to ClinVar are buried in core (not shown) or near interfaces in the predicted structure (e.g. c,d)
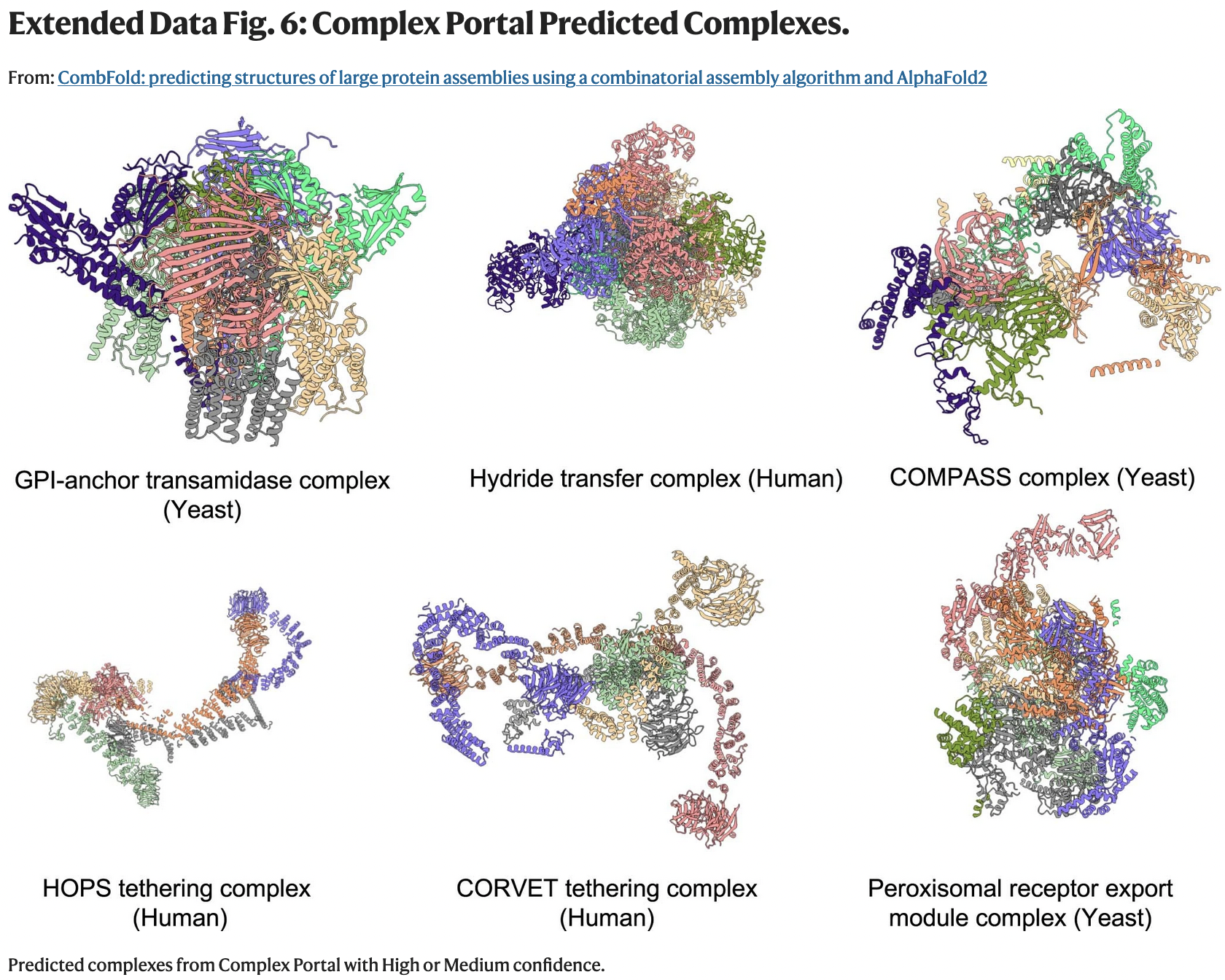
The confidence score is the weighted combination of the component pairwise PAE-based transformation scores. However, I didn't see an explanation of what values would be considered high or medium.
Stoichiometry Prediction
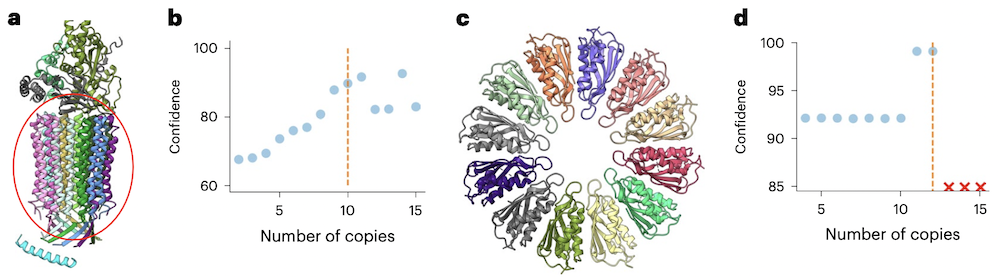
- Different stoichiometries can be enumerated using the same AFM models
as input, so that the time-consuming part (AFM) need only be run once.
Assembling the possible stoichiometries from these models is fast, and
the resulting confidence scores can be used to narrow the possibilities.
- a,b: mitochondrial ATP synthase with bound native cardiolipin, PDB
6tdx
- c,d: PelC dodecamer, PDB 5t11; 13+-mers could not be assembled without major steric clashes
CombFold Hierarachical Assembly vs. Global AFM
- Global (end-to-end) AFM is limited by maximal total residues in the complex and lack of diversity in the generated structures
- CombFold emphasizes accurately predicted pairs
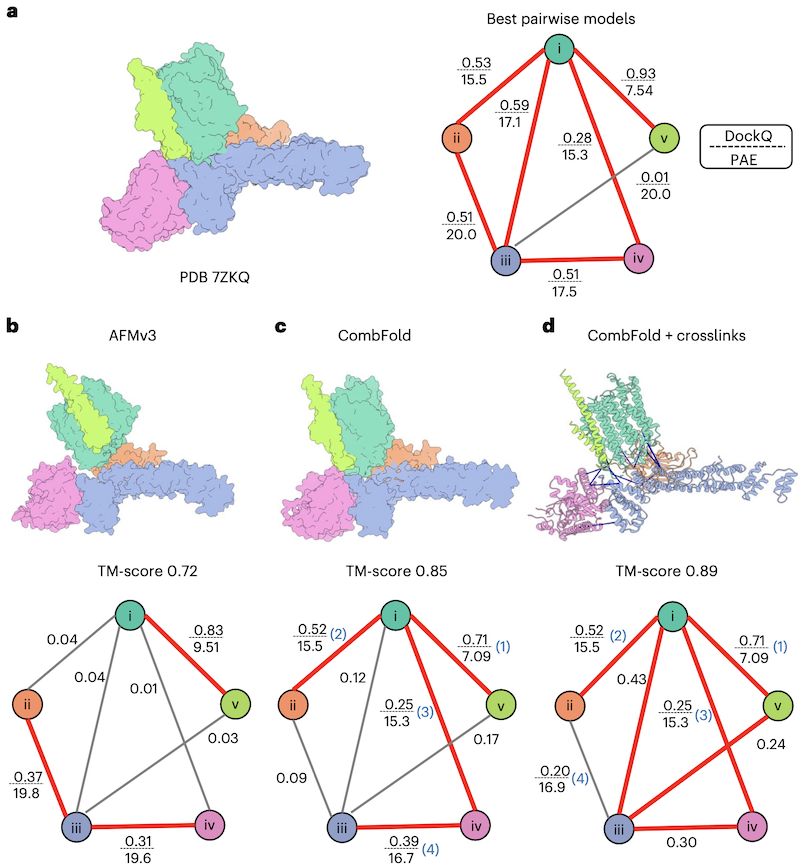
- Example: Pp module assembly intermediate of complex I
- a: CombFold finds six pairwise interactions of acceptable quality (DockQ >0.23). Several assembly pathways are possible because only four pairwise interactions that produce a spanning tree of all subunits are needed to assemble the complex. For example, see c in which assembly order is indicated with numbers in parentheses.
- b: global AFM correctly predicts only three pairwise interactions.
- d: Even if a correct pairwise interaction is not predicted by AFM (edge iii-v), it can still be generated during CombFold assembly.