AlphaFold2 vs. Crystal Structures
AlphaFold predictions: great hypotheses but no match for experiment. Terwilliger TC, Liebschner D, [...] Richardson JS, Read RJ, Adams PD. bioRxiv 2022.11.21.517405
- AlphaFold predictions can be amazingly accurate
- some have suggested that AlphaFold predictions are just as good as or even better than solved/deposited structures; they have very good stereochemistry, e.g., fewer outlier sidechain conformations than deposited structures
- the authors advise instead treating AlphaFold predictions as (high-quality)
hypotheses:
- PDB-deposited structures are in better agreement with exptl. density
- even high-confidence predictions may be inconsistent with exptl. density (and only ~40% of human proteome residues are predicted with high confidence)
- predictions differ more than the variation expected from flexibility or different crystal contacts, as represented by structures of the same protein from different crystal forms
[back to paper list]
AlphaFold vs. Experiment
- test set of 102 maps determined without reference to deposited models
- ...with resulting deposited structures determined at free R 0.30 or better
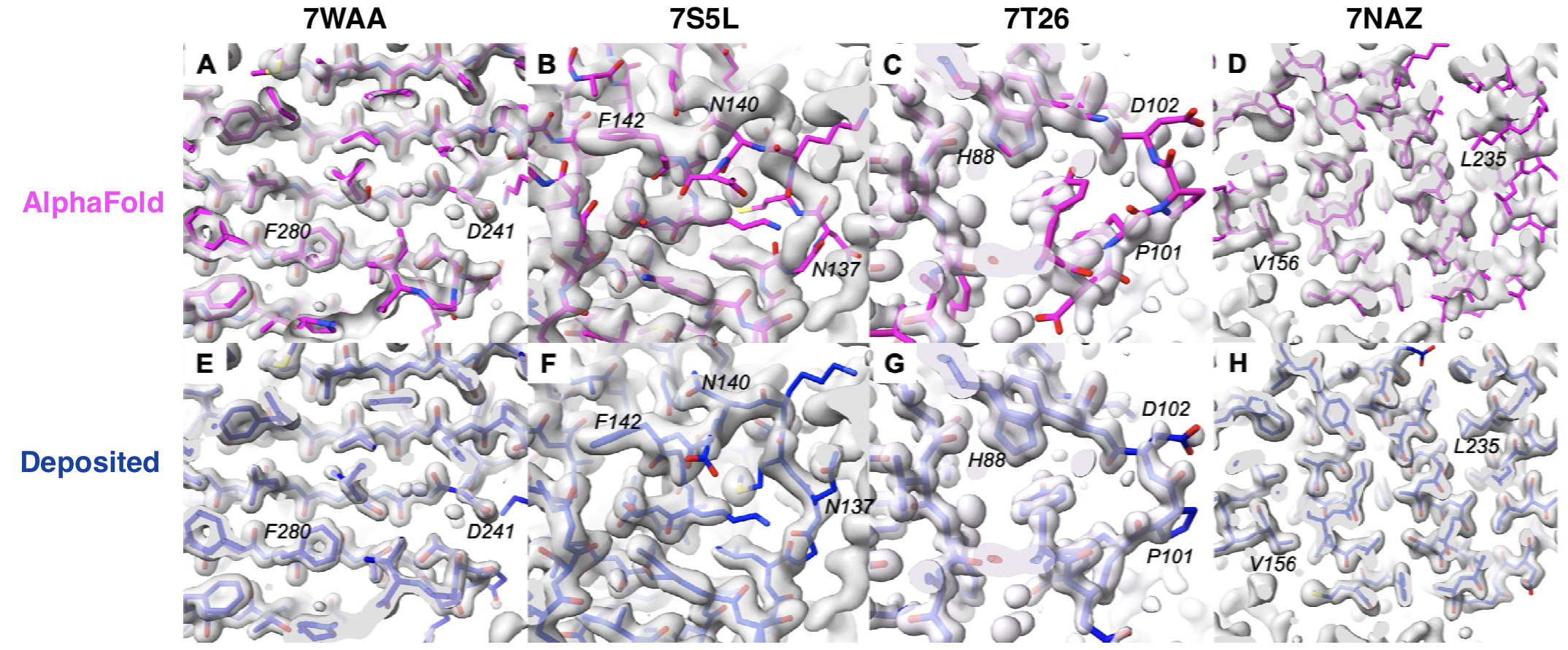
- all residues in the figure are predicted by AlphaFold at high confidence (pLDDT >90) for density maps with resolution 1.1-1.6 Å contoured at 1.1-1.9 σ
- 7WAA: highly accurate prediction (map-model correlation when superimposed on deposited structure 0.72, Cα RMSD vs. deposited 0.5 Å)
- 7S5L: areas not consistent with density (correlation 0.44, Cα RMSD 2.1 Å)
- 7T26: not consistent with density but plausible altloc where density is weaker
- 7NAZ: prediction distorted relative to map
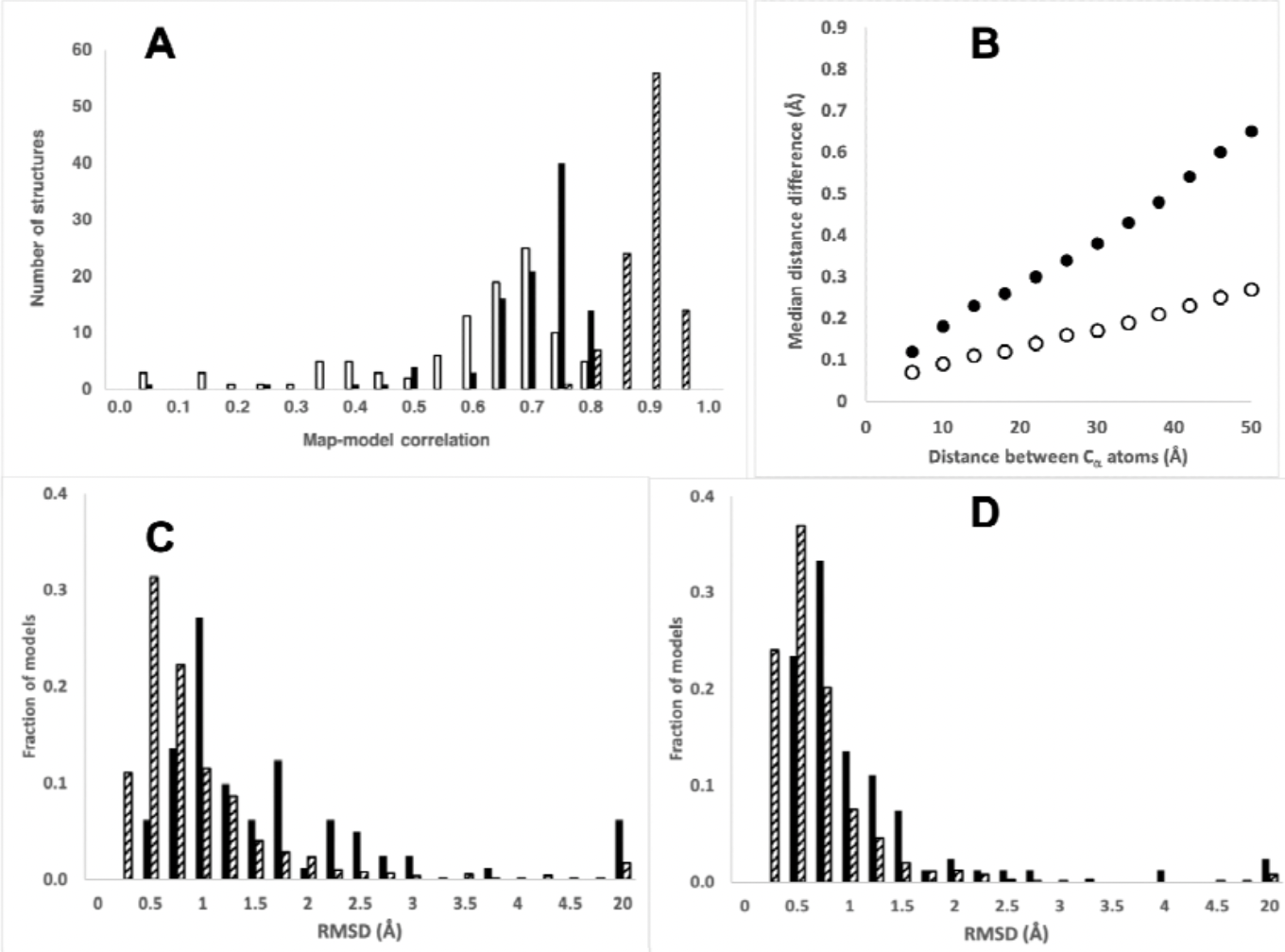
Overall Statistics for the 102 Predictions
- A. Map-model correlation: AlphaFold (open, mean 0.56), deposited (hatched, 0.86), AlphaFold “morphed” (solid, 0.67)
- B. Median difference in intramodel Cα-Cα distances: AlphaFold moderate-to-high confidence parts (pLDDT>70) vs. deposited (solid), 926 pairs of high-resolution structures from different crystal forms/space groups (open)
- C. Cα RMSD: AlphaFold vs. deposited (solid, median 1.0 Å), pairs from different crystal forms (hatched, 0.6 Å)
- D. As in C, except “morphed” with smoothed distortion field to enhance global similarity while retaining local differences (median RMSD for both now 0.4 Å)
- ∴ AlphaFold discrepancies are mostly from distortions rather than alternative domain packing
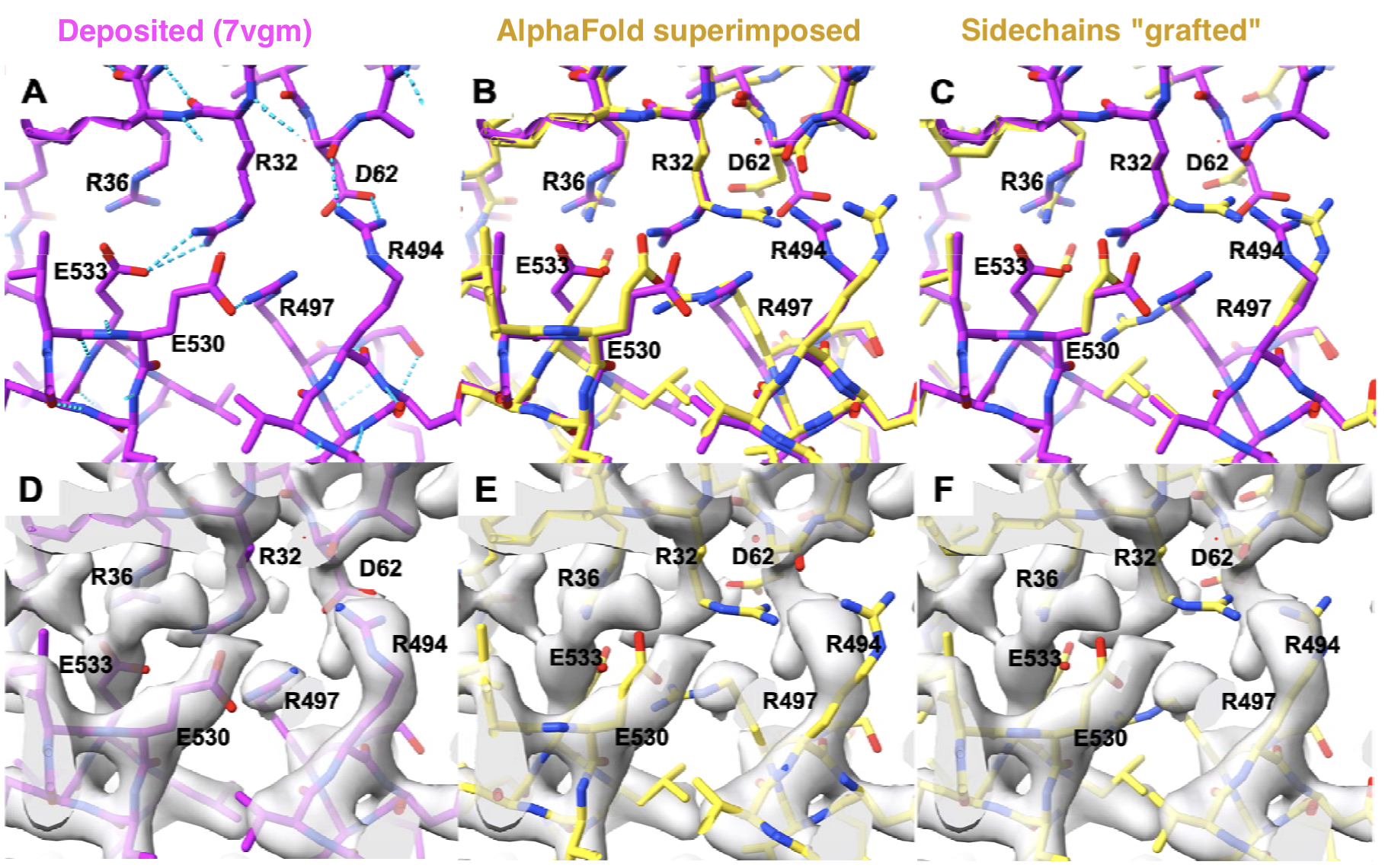
Focusing on the Sidechains
- grafting: superimposing each residue separately using its backbone atoms
- 7vgm density-map resolution: 2.3 Å
- for the dataset of 102 predictions, 20% of sidechains in residues with moderate-high pLDDT also had RMSDs vs. deposited (beyond Cβ) ≥ 1.5 Å, and 7% were clearly incompatible with the density (p < 0.01 based on the uncertainty of the density and the number of sidechain atoms)
- the proportion was about the same for functionally important residues (7% incompatible)
- comparing exptl. but from different space groups, 6% with RMSD ≥ 1.5 Å, 2% incompatible with density
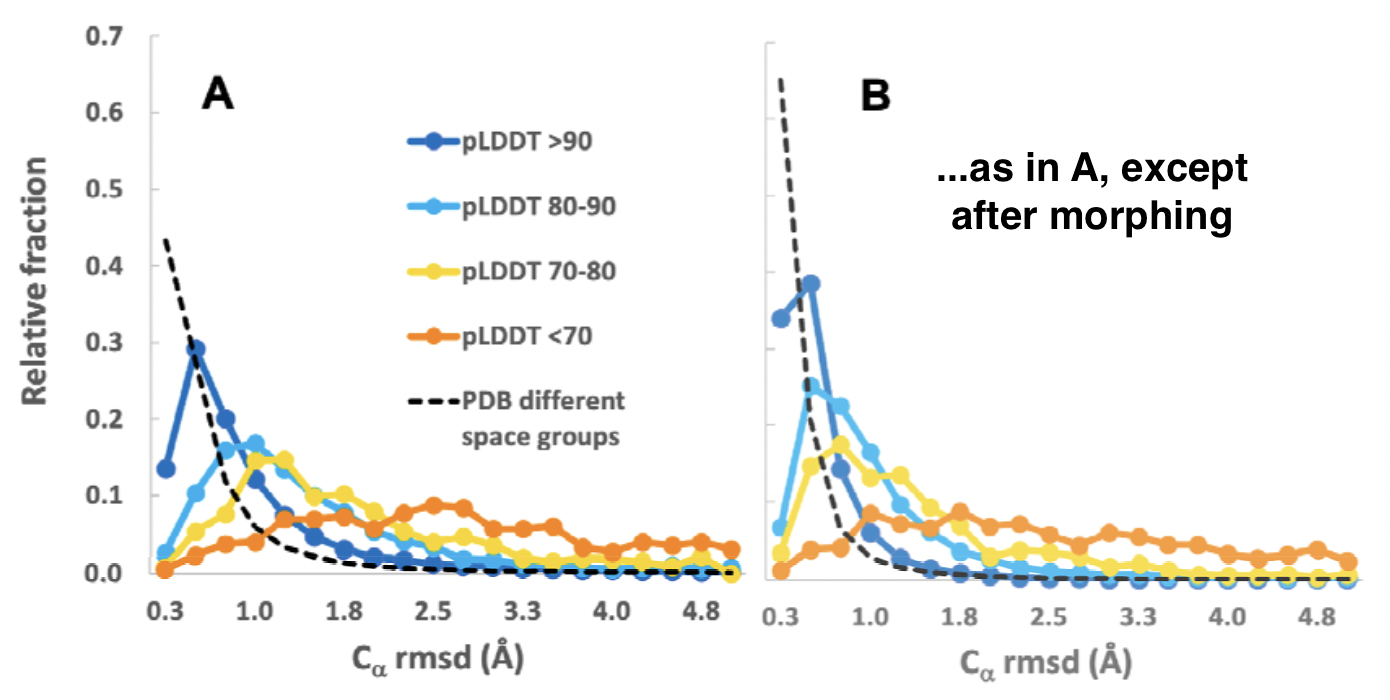
Cα Error (Cα-Cα Distance) after Superposition
- smaller for higher-confidence parts, but larger than one might expect, with long tails to higher values
| AlphaFold confidence (pLDDT) | Median error (Å) | % > 2 Å |
|---|---|---|
| >90 | 0.6 | 10 |
| 80-90 | 1.1 | 22 |
| 70-80 | 1.5 | 33 |
| <70 | 3.5 | 77 |
- for diff-space-group pairs: median Cα-Cα distance 0.3 Å, 5% > 2 Å
Limitations and Controls
- Is sidechain grafting unfair where the mainchain conformation
is different?
- Limiting the comparison to positions with similar mainchain angles gave much the same results (~17-18% different orientation, 7% clearly incompatible)
- Is the test set (chosen for high-quality data) too small, with too few
residues predicted at low confidence?
- a larger-scale comparison by DeepMind found higher median Cα RMSD (2.3 Å, 1.5 Å excluding the worst 5%, as compared to 1.0 Å in Fig 2C, slide 3), so this study was conservative in its approach
- AlphaFold multimer was not used, leading to large errors in cases of
domain-swapping or other significant rearrangements due to multimerization
- Median values were used for comparison to prevent overweighting of worst cases
- AlphaFold was used without templates
- Comparison of 81 models from the AlphaFold database (made with templates) vs. deposited gave the same median Cα RMSD, 1.15 Å, as the 81 models (made without templates) from the test set of 102
Combining AlphaFold & Experimental Data
Improved AlphaFold modeling with implicit experimental information.
Terwilliger TC, Poon BK, Afonine PV, Schlicksup CJ, Croll TI, Millán C, Richardson JS, Read RJ, Adams PD.
Nat Methods. 2022 Nov;19(11):1376-1382.
(Phenix version of AlphaFold2 Colab:
https://github.com/phenix-project/Colabs)
Accelerating crystal structure determination with iterative AlphaFold prediction. Terwilliger TC, Afonine PV, Liebschner D, Croll TI, McCoy AJ, Oeffner RD, Williams CJ, Poon BK, Richardson JS, Read RJ, Adams PD. Acta Crystallogr D Struct Biol. 2023 Mar 1;79(Pt 3):234-244.