flotillin-surfs-portrait.cxs
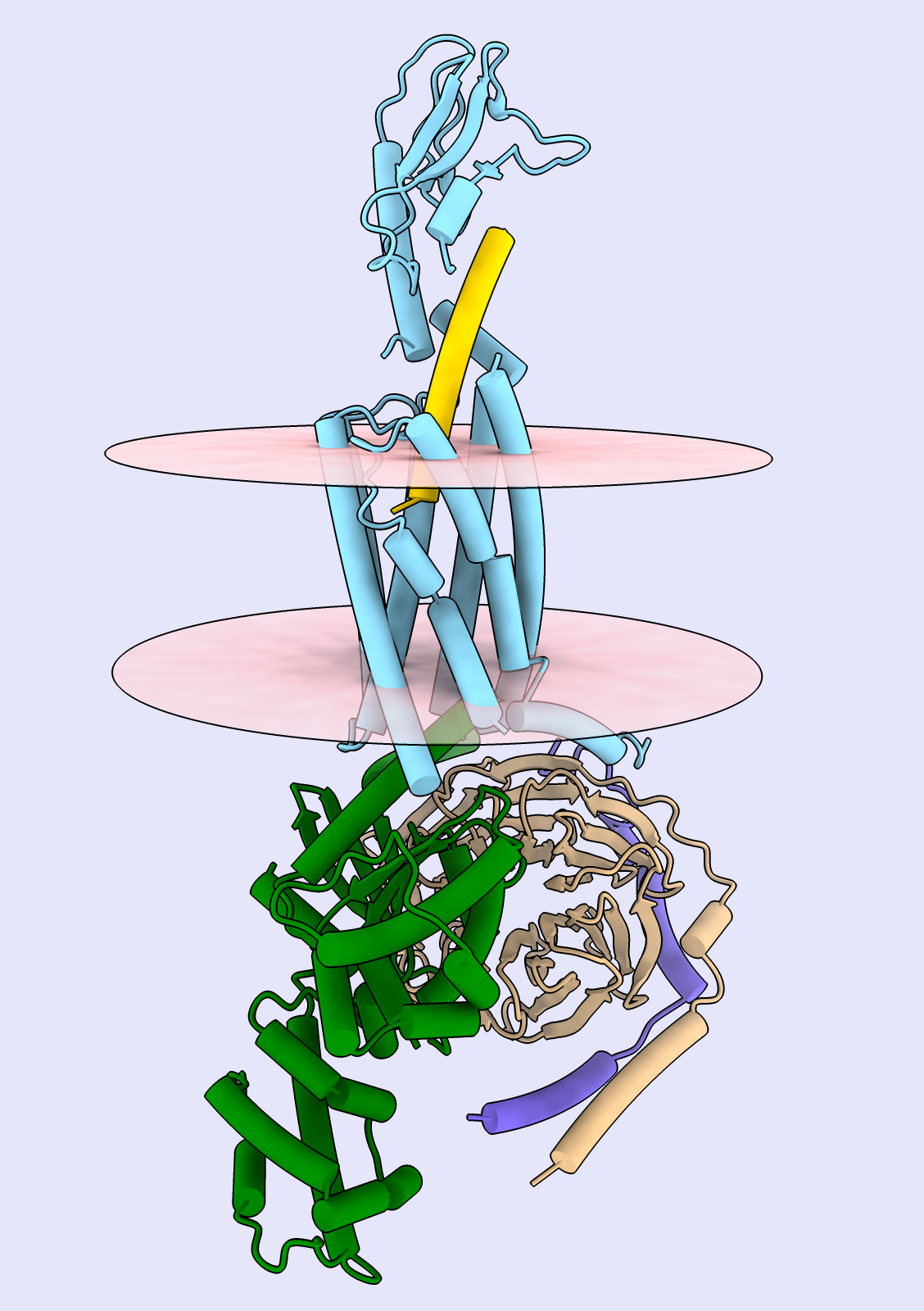
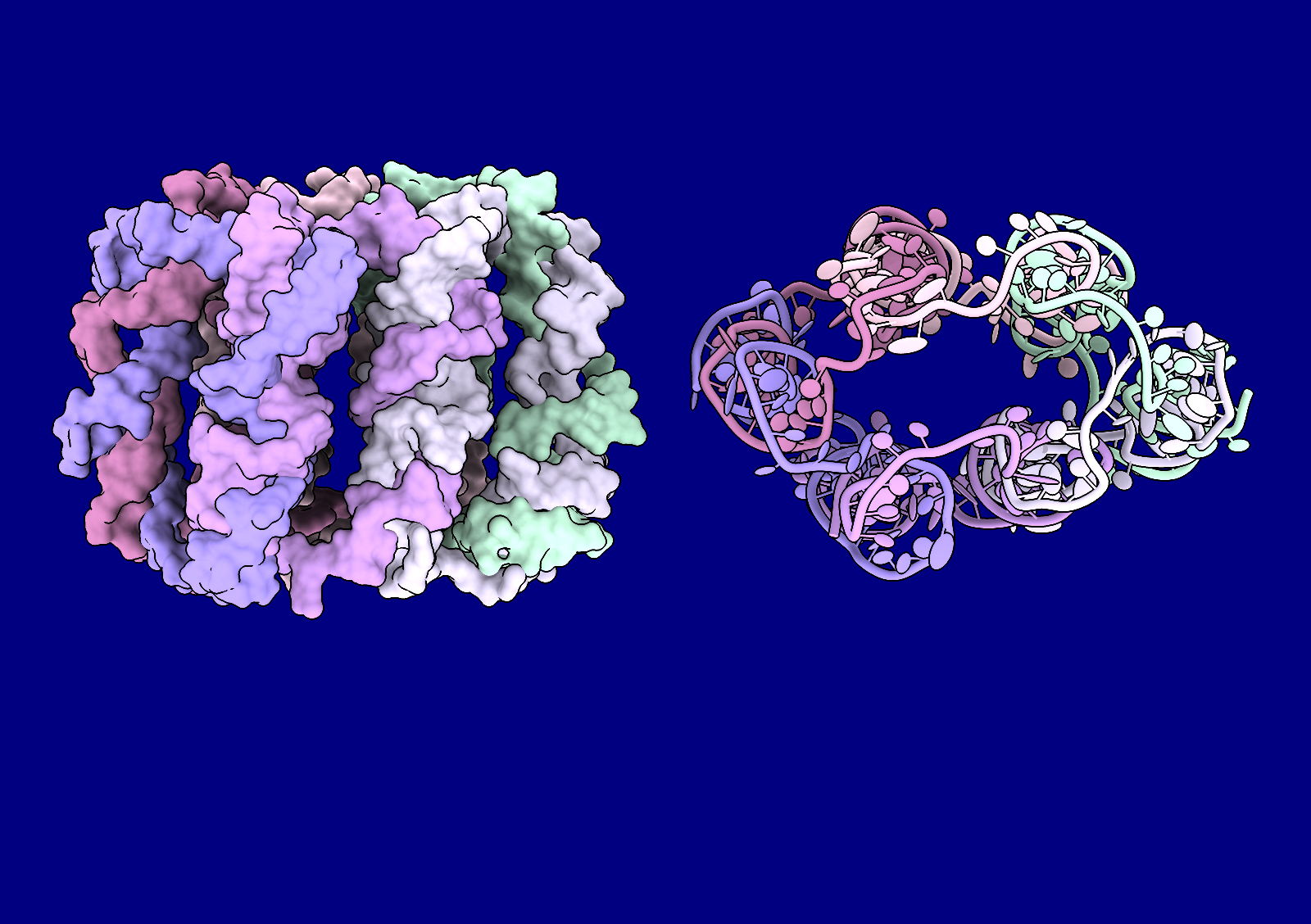
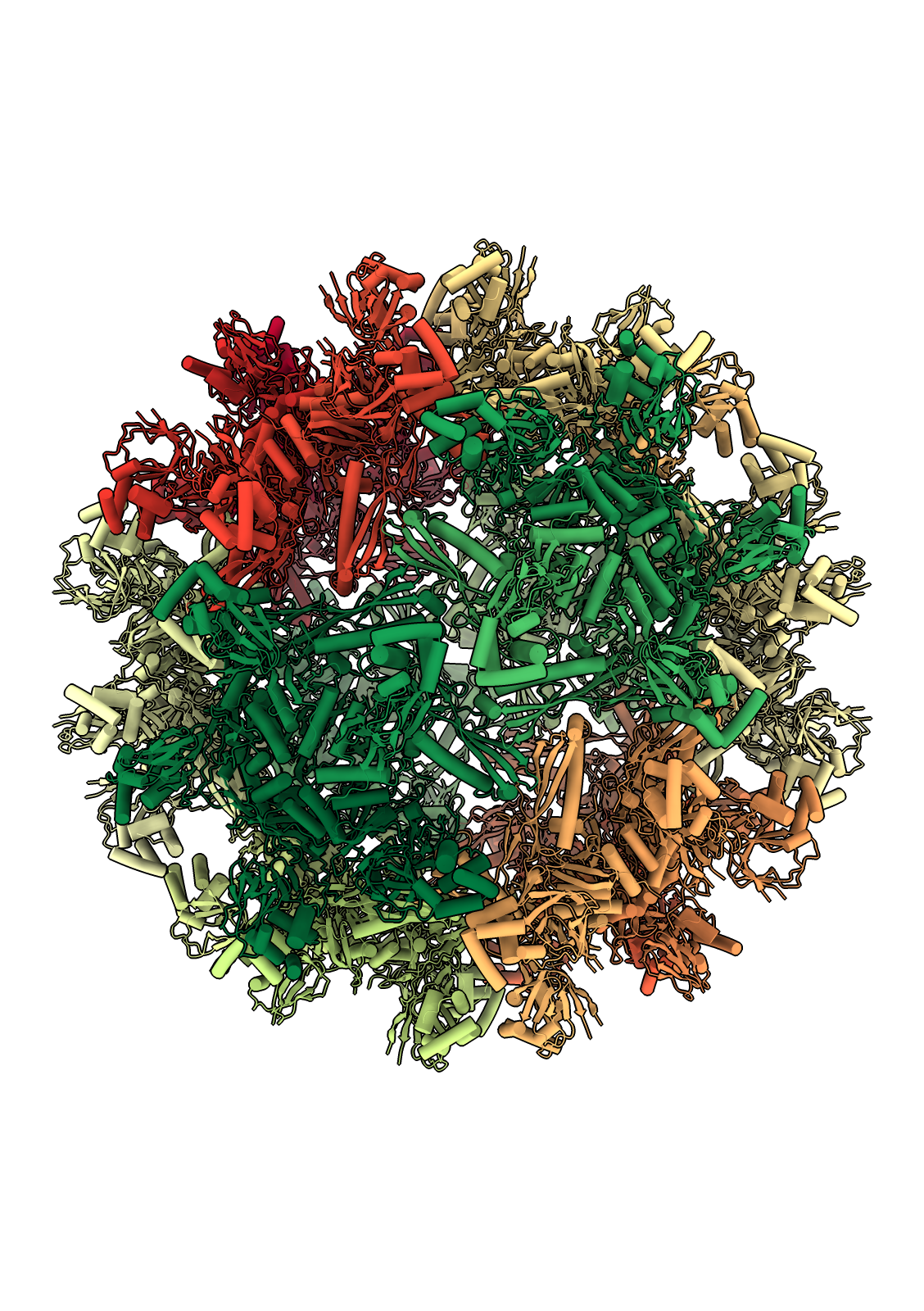
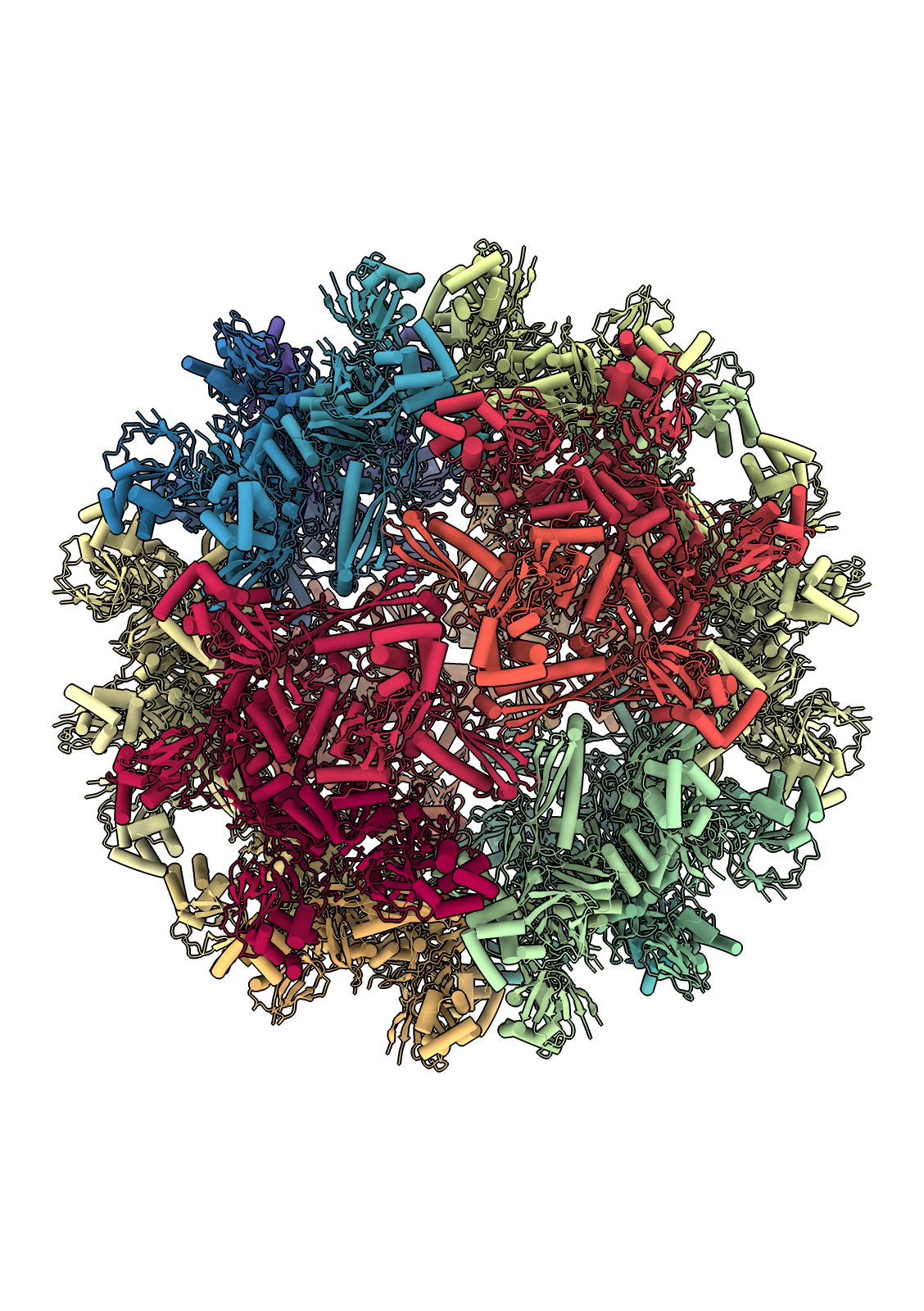
The flotillin complex is involved in membrane trafficking and the formation of membrane microdomains in mammalian cells. It has a tapered barrel or flowerpot shape formed by alternating flotillin-1 (here in green) and flotillin-2 (red) subunits, where both the wide and narrow ends may form contacts with the lipid membranes. The flowerpot rim is “decorated” with cysteine sulfur atoms (yellow). This structure is human flotillin from PDB entry 9BQ2 (Fu and McKinnon, Proc Natl Acad Sci 121:e2409334121 (2024)). [camera ortho, sym assembly 1 copies false, bg steel blue, /A indianred, /B green, gold :cys&S with atomrad 2.5, full lighting except depthcue off]