
Tool: Orthopick
Orthopick contains settings for the
mouse mode named
orthopick,
developed for picking particles in cryo-electron tomograms with the mouse.
The orthopick mode shows three orthogonal slices of a small area of
the map centered at the clicked point as well as laid out in a plane
for easy viewing without rotation.
For an example of use, details, and discussion, see
Membrane Complexes. See also:
Volume Viewer
Orthopick can be started
from the Volume Data section of the Tools menu and
moved, resized, etc.
like other tool windows. Starting Orthopick automatically
assigns the mode to the right mouse button
(= trackpad + Alt on Windows, trackpad +
 on Mac).
Other ways to assign the mode are by clicking the orthopick icon
on Mac).
Other ways to assign the mode are by clicking the orthopick icon
 in the Toolbar or by using the
mousemode command.
By default, the orthopick icon
in the Toolbar or by using the
mousemode command.
By default, the orthopick icon
 is
not shown in the Toolbar,
but it can be added to the Home tab using the
Toolbar Settings.
is
not shown in the Toolbar,
but it can be added to the Home tab using the
Toolbar Settings.
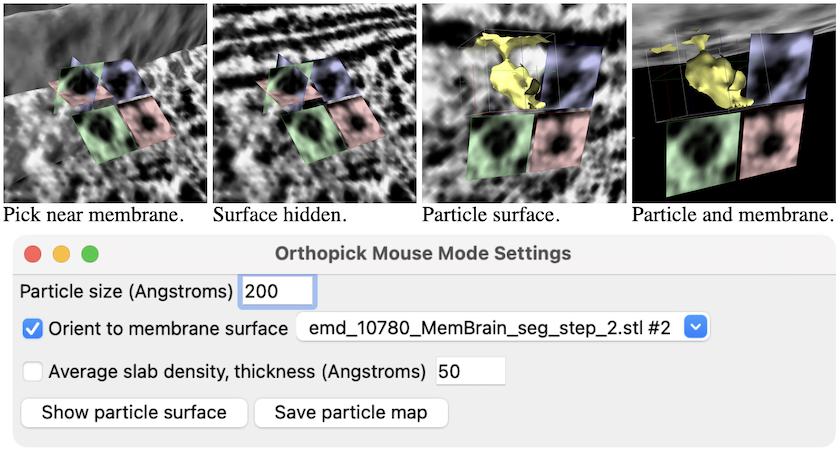
In the orthopick mode,
- clicking shows orthogonal red (cryo-EM-beam-aligned XY), green (XZ),
and blue (YZ) slices centered on the click, as well as copies of the
slices laid out in a plane, as shown in the figure
- each of the slice planes can be dragged to see different cross-sections
- dragging with the Shift key held down translates (recenters) the box
delimiting the orthoplanes
Settings:
- Particle size (Angstroms) [default 200]
– orthoplanes box size in each dimension
- Orient to membrane surface [surface-model]
– if a surface model is specified, use the plane of the surface
as the reference for aligning the box; if no surface model is specified,
the box will be aligned to the XYZ axes of the tomogram
- Average slab density, thickness (Angstroms) [default 50]
– color the slice planes based on average density values through a
slab of the specified thickness
Buttons:
- Show particle surface –
show 3D contour surface inside the box instead of intersecting orthoplanes;
the slices laid out in a plane will still be shown, however, as in
the righthand image above. The contour level can be changed by
clicking and dragging with the orthopick-assigned mouse button
on the particle surface.
- Save particle map –
save an MRC file of the map inside the box, including the origin
and rotation to apply if the file is opened in ChimeraX;
other programs may honor the origin, but not the rotational information,
as the latter is not a defined part of the MRC format
- Help – show this page
in the Help Viewer
UCSF Resource for Biocomputing, Visualization, and Informatics /
August 2025

![]() on Mac).
Other ways to assign the mode are by clicking the orthopick icon
on Mac).
Other ways to assign the mode are by clicking the orthopick icon
![]() in the Toolbar or by using the
mousemode command.
By default, the orthopick icon
in the Toolbar or by using the
mousemode command.
By default, the orthopick icon
![]() is
not shown in the Toolbar,
but it can be added to the Home tab using the
Toolbar Settings.
is
not shown in the Toolbar,
but it can be added to the Home tab using the
Toolbar Settings.