TALAIA: simplified 3D protein representations
August 8, 2023
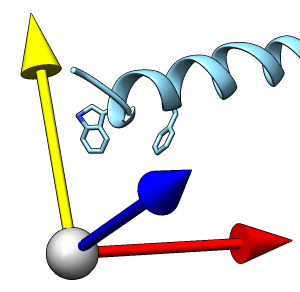
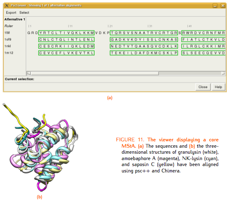
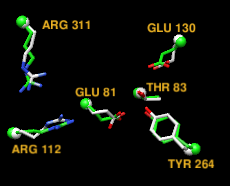
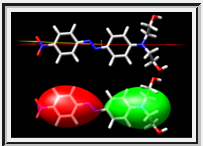
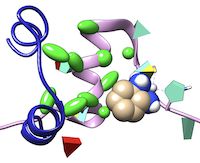
TALAIA represents different types of amino acid residues as simplified objects with different shapes and colors, to facilitate rapid recognition of molecular properties and types of interactions. TALAIA is described in Alemany-Chavarria et al., Bioinformatics 39:btad476 (2023).