
Tom Goddard
July 28, 2005
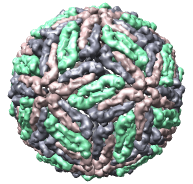
We'll look at one of the viruses that cause the common cold, Human Rhinovirus 2, Protein Data Bank structure 1v9u. This tutorial is based on Chimera version 1.2129, but should also work on earlier versions of Chimera.

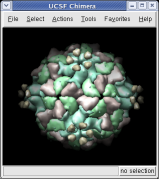
With a three button mouse drag the right mouse button in the graphics window to zoom in and out. The middle button lets you move the position of the virus capsid model, and the left mouse button rotates it.
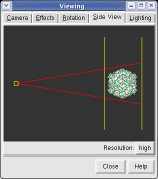
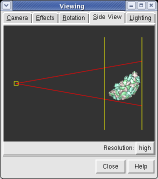
The far portion of the capsid is invisible. Use menu entry Favorites / Side View and drag the right vertical yellow line further to the right. The two yellow lines are near and far clipping planes and the yellow square represents your eye position in this view from the side.
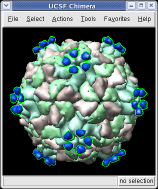
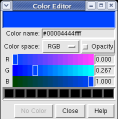
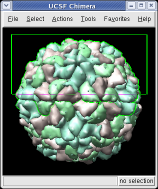
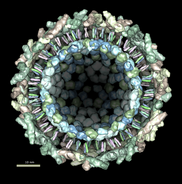
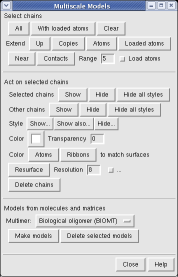
This structure shows part of the receptor found on the surface of cells in your nose that the virus recognizes. These are the chains that appear in rings of 5 near the 5-fold symmetry axes. We'll color the receptor so it stands out. Click with Ctrl left mouse button on one of the receptor surfaces to select it. You need to hold down the Ctrl key on the keyboard when selecting objects. The selected surface is shown with a green outline. Then press the Copies button on the second line of the Multiscale dialog. Then press the Color button in the middle of the dialog. It has a raised border with the color shown in the middle. Then change the color using the color dialog.
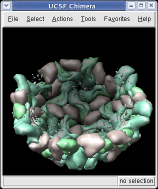
Press the Hide button in the Multiscale dialog on the line that says Selected chains to hide the receptors. To see the inside of the capsid, select half of the capsid by dragging a rectangle using Ctrl left mouse button, then press the Hide button and rotate the model with the left mouse button (without holding down the ctrl key) to see the interior of the capsid.
Chimera makes objects that are further from view dimmer. Adjusting the clip planes by moving the vertical yellow lines in the side view dialog (menu entry Favorites / Side View) changes the distance at which the dimming effect starts and ends.

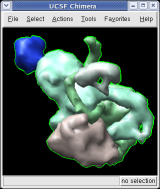

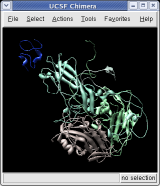
To show the asymmetric unit, that is 1/60th of the icosahedral capsid, click the With loaded atoms button at the top of the Multiscale dialog, then press the Show button on the line saying "Selected chains" and the Hide button on the line in the dialog that says "Other chains". Some of the tiny blobs shown are parts of larger proteins. Change the Resolution field in the dialog from 8 angstroms to 4 and press the Resurface button. The green protein has two arms that wrap around the light blue protein.
On the Style line of the dialog press the Show... button which will display a menu, and select Ball and Stick to show all atoms of the selected proteins. Press the Color Atoms button to make the atoms colors match the original surface depiction colors. Similarly you can display just a ribbon of the protein backbones by choosing Ribbon style and pressing Color Ribbons to make the ribbon colors match the original surface protein colors.
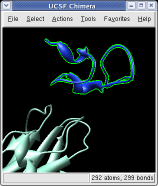
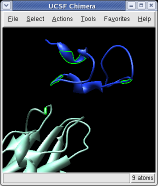
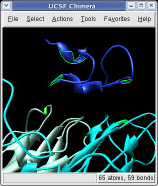
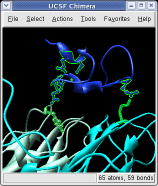
Ctrl left click on the blue receptor ribbon. That selects one residue. Then press the up arrow key on the keyboard to promote the selection to the whole ribbon. Change the Range value near the top of the Multiscale dialog from 5 angstroms to 3, turn on the load atoms switch, and press Contacts. This finds all residues within 3 angstroms of the receptor. Some contacts are with an unshown protein. Use Style Show... and choose Ribbon to show the other contacting protein. To show the atoms involved in the contacts use the Actions menu at the top of the graphics window, Actions / Atoms+Bonds / show. To show all atoms in the contacting residues press the up arrow key on the keyboard to exted the selection to the containing residues and use Actions / Atoms+Bonds / show again.

| 
| 
| 
|
| Reovirus core, 1ej6 | Dengue virus, 1k4r | Ribgrass mosaic virus (helical), 1rmv | Sindbis virus, 1ld4 |
There are about 200 virus capsid structures known at atomic resolution (ca. 2005). Below are some of the Protein Data Bank identifiers. The Virus Particle Explorer web site contains information about these virus structures.
1a34 1a6c 1al0 1al2 1aq3 1aq4 1ar6 1ar7 1ar8 1ar9 1asj 1auy 1aym 1ayn 1b35 1bbt 1bev 1bms 1bmv 1c8d 1c8e 1c8f 1c8g 1c8h 1c8m 1c8n 1cd3 1cov 1cwp 1d3e 1d3i 1d4m 1ddl 1dgi 1dnv 1dwn 1dyl 1dzl 1dzs 1e57 1e6t 1e7x 1eah 1ej6 1ev1 1f15 1f2n 1f8v 1fmd 1fod 1fpn 1fpv 1fr5 1frs 1gav 1gff 1gkv 1gkw 1gw7 1gw8 1h8j 1h8t 1hb5 1hb7 1hb9 1hdw 1he0 1he6 1hri 1hrv 1hxs 1if0 1ihm 1ijs 1jew 1js9 1k3v 1k4r 1k5m 1kuo 1kvp 1laj 1ld4 1lp3 1m06 1m0f 1m11 1m1c 1m4x 1mec 1mqt 1mst 1mva 1mvb 1mvm 1n6g 1na1 1na4 1ncq 1ncr 1nd2 1nd3 1ng0 1nn8 1nov 1ny7 1ohf 1ohg 1oop 1opo 1p58 1p5w 1p5y 1pgl 1pgw 1piv 1po1 1po2 1pov 1pvc 1qbe 1qgc 1qgt 1qju 1qjx 1qjy 1qjz 1qqp 1r08 1r09 1r1a 1rb8 1rhi 1rmu 1ruc 1rud 1rue 1ruf 1rug 1ruh 1rui 1ruj 1rvf 1s58 1sid 1sie 1smv 1stm 1sva 1tge 1thd 1tme 1tmf 1tnv 1u1y 1uf2 1upn 1v9u 1vak 1vb2 1vb4 1vba 1vbb 1vbc 1vbd 1vbe 1vrh 1w39 1wcd 1wce 1x9p 1x9t 1yc6 1zdh 1zdi 1zdj 1zdk 2bbv 2bpa 2btv 2cas 2hwb 2hwc 2hwd 2hwe 2hwf 2mev 2ms2 2plv 2r04 2r06 2r07 2rm2 2rmu 2rr1 2rs1 2rs3 2rs5 2stv 2tbv 4dpv 4rhv 4sbv 5msf 6msf 7msf
Virus structures with only alpha-carbon coordinates. Some have atomic coordinates for only the alpha-carbon atoms. An example is Protein Data Bank model 1d3e, Human Rhinovirus 16. To display these using the multiscale tool you must adjust the surfacing parameters before the pressing the Make models button. Click the checkbox at the end of the Resurface line and adjust Threshold atom density from 0.02 to 0.002 for these viruses. Without this change no surface will appear.
Virus structures without biological unit matrices. About half of the virus models from the Protein Data Bank (primarily older ones) do not contain the symmetry matrices needed to generate the capsid. When you press the Make models button you just get surfaces for the asymmetric unit. For these viruses you can download alternate coordinates from the Virus Particle Explorer web site. Then set the Multimer type to Icosahedral symmetry, ViPER Z(2)35X(2), before pressing Make models.
Chimera
Other Sites