EM Data Visualization with Chimera
Tom Goddard
December 1, 2005
Introduction to Chimera
- Chimera is an interactive molecular graphics program.
- Will show EM density map and large model display capabilities.
- Most Chimera capabilities are for studying single macromolecules.
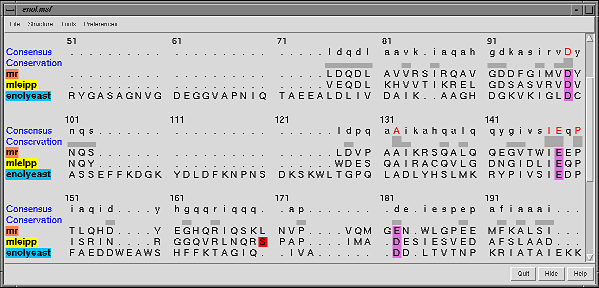
- Multiple sequence alignment features are particularly strong.
Chimera Development
- Chimera developed by the
Resource for Biocomputing, Visualization and Informatics at
UC San Francisco, funded by NIH.
- Currently have 5 developers.
- Available on Windows, Mac, Linux, free for academic use.
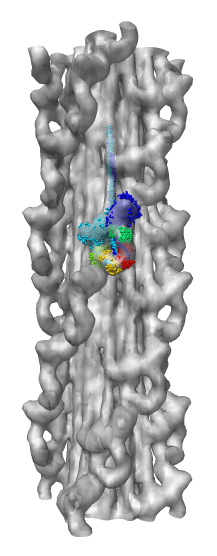
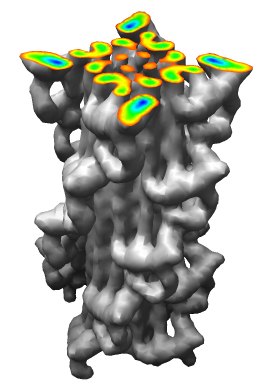
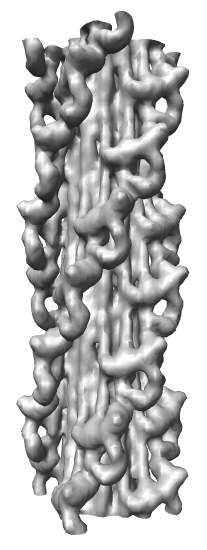
Demonstration Showing Myosin Thick Filament
Map and model fitting provided by Roger Craig's lab at
Univ. of Massachusetts Medical School. Published in Nature, August 2005.
Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padron R.
Atomic model of a myosin filament in the relaxed state.
Nature. 2005 Aug 25;436(7054):1195-9.
Significance: Their map has a factor of 2 better resolution
than previous myosin thick filament data and for the first time allows
unambiguous fitting of myosin atomic models. The fitting reveals
intermolecular contacts that may be important for maintaining the
relaxed muscle state.
Chimera Web Page
- Chimera web site (Google: UCSF Chimera),
http://www.cgl.ucsf.edu/chimera.
- Production releases about once per year.
- Snapshot releases about once a month include recent features.
- Extensive documentation.
Web site offline.
Show download page, image gallery, guide to volume data display.
Demo Outline
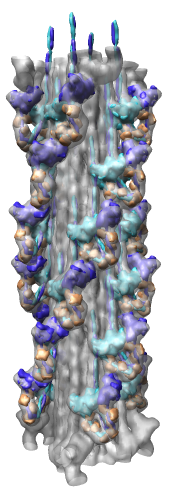
Map Details
- Myosin thick filament from striated tarantula muscle.
- Single particle cryo-EM reconstruction.
- Reconstruction used ~60 filaments, ~7200 pairs of myosin heads.
- 2.5 nm resolution.
- Relaxed muscle state where myosin is not bound to nearby actin filaments.
- Two symmetries: 1) 4-fold symmetry about axis (rotation by 90 degrees),
and 2) helical symmetry, rotate by 30 degrees, shift by 14.5 nm.
- About 35 nm diameter.
|
|

|
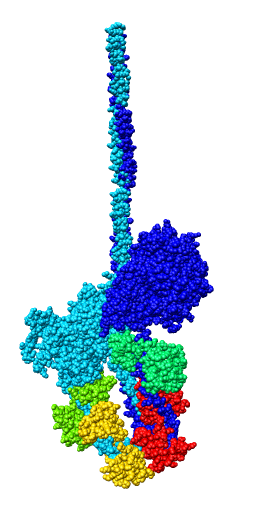
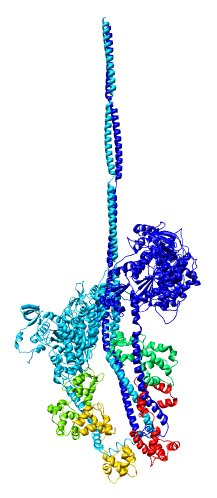
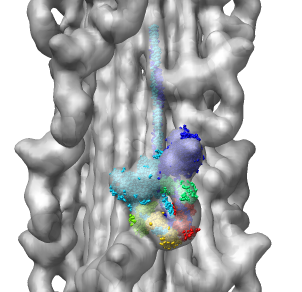
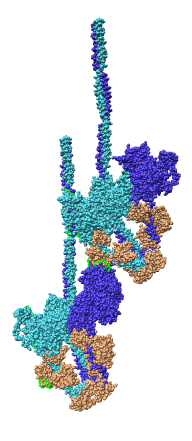
Model Details
- Asymmetric unit contains 6 chains: 2 myosin heavy chains (blue, cyan),
2 essential light chains (green), 2 regulatory light chains (yellow, red).
- Each heavy chain forms myosin head which is the actin binding motor domain,
and a long alpha helix tail.
- Motor domains walk along nearby actin filaments in muscle tissue.
- Alpha helix tails of two heavy chains form a single coiled coil tail.
- Tail is ~6 times longer than depicted (~180 nm).
- Light chains wrap around heavy chain alpha helices.
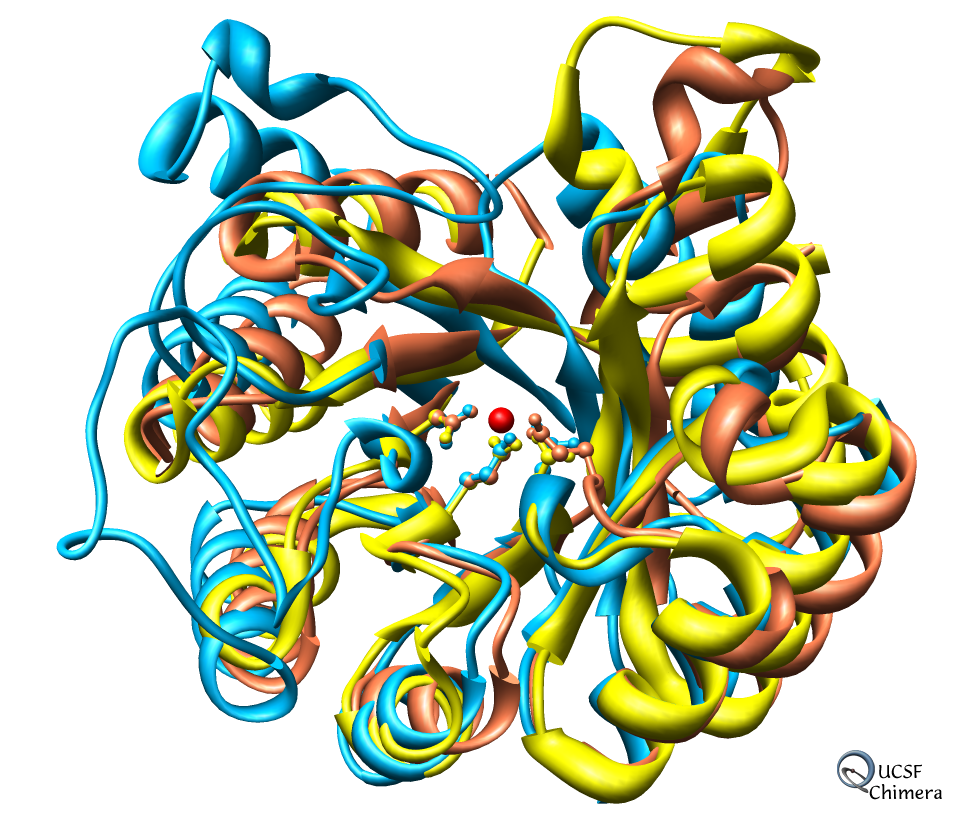
- Model is from hand fitting chicken gizzard smooth muscle (PDB 1I84) to map
as 4 separate rigid pieces.
- 1I84 is from fitting crystal structures 1MYS and 1BR1 into 20 angstrom
resolution cryo-EM map from tilt series of 2D-crystals.
|
|
|
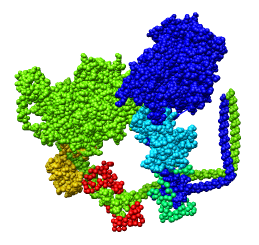
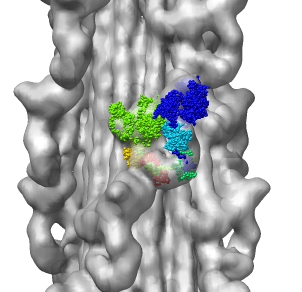
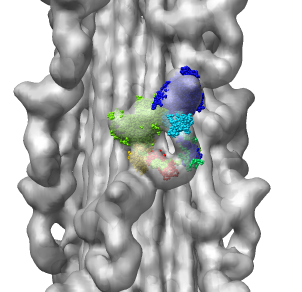
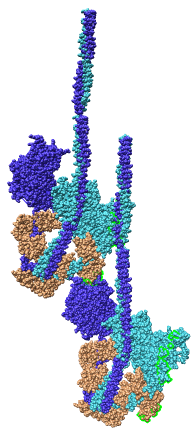
Fitting Atomic Model in Map
Fit 1i84 into map. Hand place model approximately in map. Use
Tools -> Volume Data -> Fit Models in Maps to optimize model
position in map. First hide incorrectly oriented tail using two
drag selections. Then select all visible atoms for fit optimization.
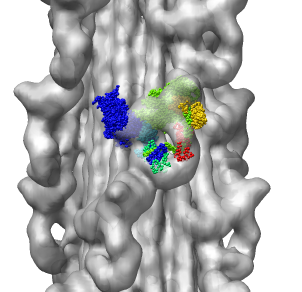
Correctly oriented fit gives density 0.84 per atom. Can hand orient
backwards getting poor fit, average density 0.61. Explain that
Woodhead, et al. broke 1i84 into 4 rigid pieces fit separately by hand.
| 
| 
|
| 1i84
| Hand placed in map.
| Locally optimized.
|
| 
|
| Hand placed backwards and optimized.
| Model from Woodhead, et al.
|
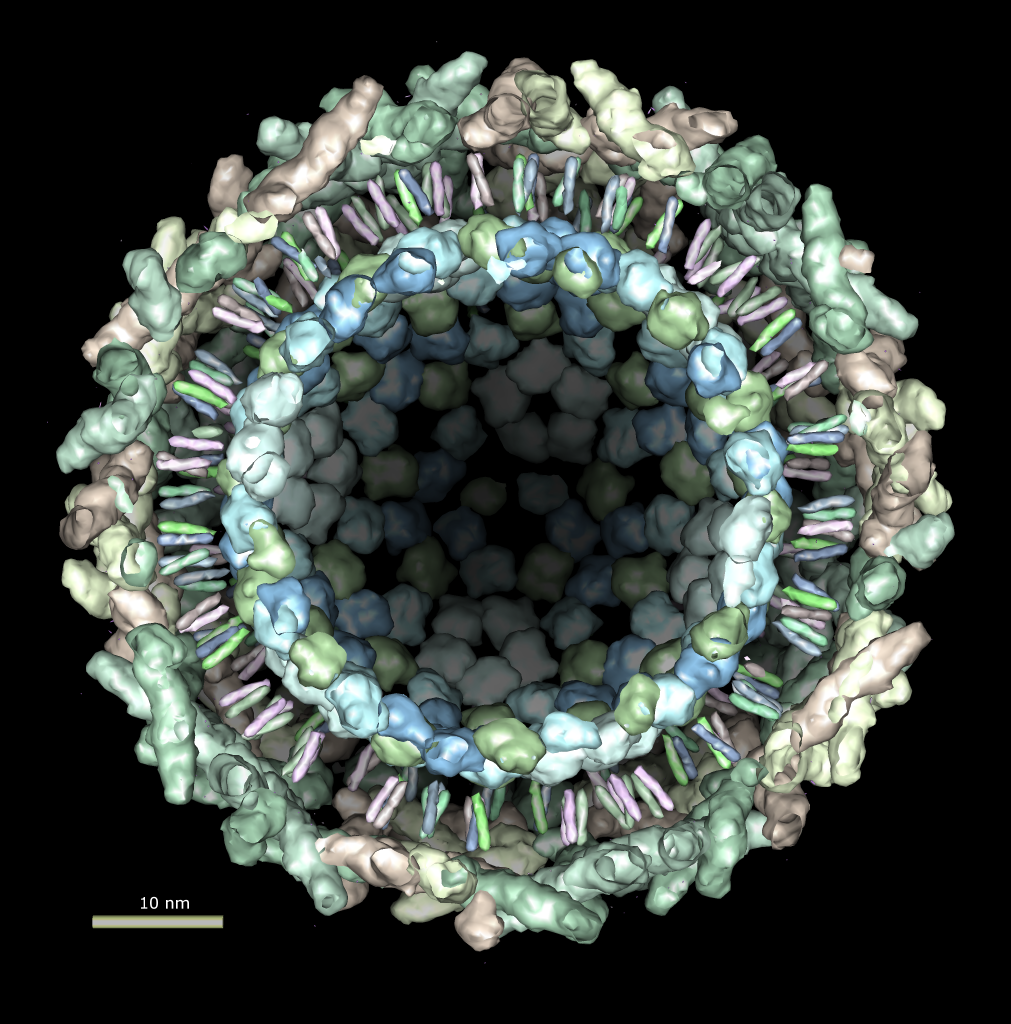
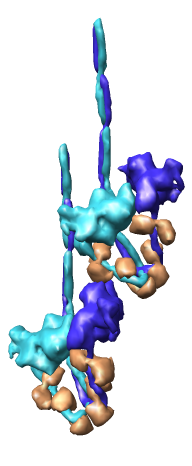
Multiscale Tool and Contacts
I placed matrices in PDB header specifying helical symmetry.
Multiscale tool reads those matrices to show 24 copies of asymmetric unit.
Switched to 6A multiscale resolution, and changed colors by hand.
Contacts between two adjacent units may be significant for holding the
myosin heads against the filament core preventing binding to actin.
Tail of lower unit contacts motor domain of upper unit. Motor domain
of lower unit conacts light chain of upper unit.
Tails are actually ~6 times longer than depicted, reaching 3 or 4 additional
myosin heads along the thick filament. The 3 or 4 tails packed in each
of the 12 subfilaments has not been modelled by Woodhead, et al.
Main point: Multiscale is useful for looking at contacts between copies
of asymmetric unit.
| 
| 
| 
|
| 24 copies of myosin pairs shown as low resolution surfaces.
| Two contacting asymmetric units.
| Contact areas highlighted in green.
| View from inside thick filament.
|
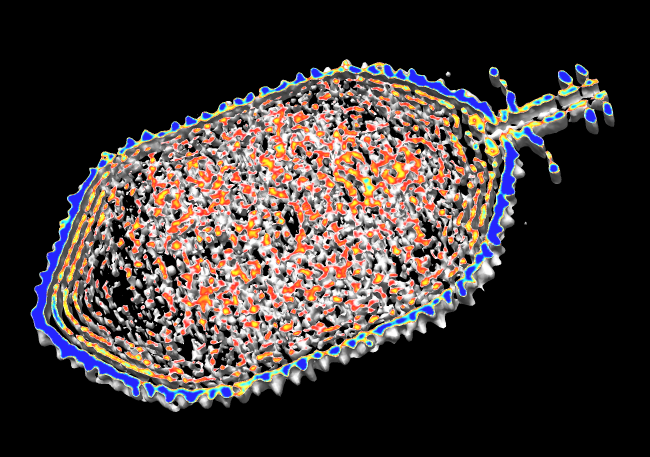
EM Map Visualization Capabilities
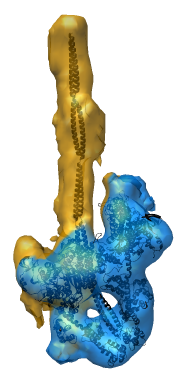
Radial Coloring
- Color twelve subfilaments that make up core of myosin thick filament
distinctly.
- Radially color map using 2 colors, orange <= 120 A, blue >= 121 A.
- Make blue invisible using 100% transparency. Use one-sided lighting
surface option in volume viewer to make holes dark.
|
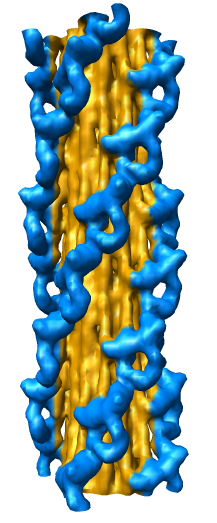
|

|
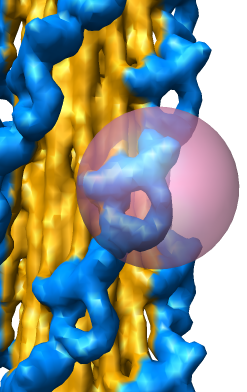
Subregion Extraction
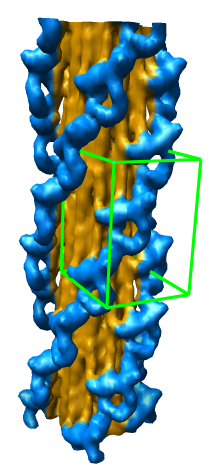
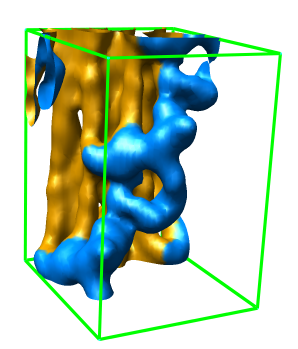
Zone Extraction
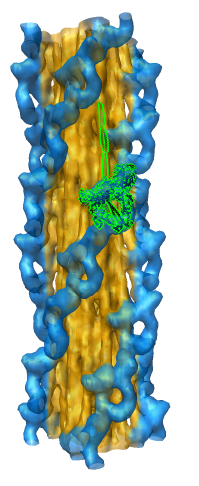
Eraser Sphere
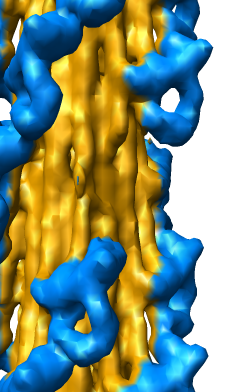
Slicing, Capping, Coloring, and Movie