Tom Goddard
December 15, 2008
In 2001, NVIDIA was first to produce a commodity chip capable of programmable shading, the GeForce 3 (code named NV20).
In 2002, the ATI Radeon 9700 (also known as R300) pixel and vertex shaders could implement looping and lengthy floating point math.
Earlier cards provided "fixed function" capabilities -- predefined lighting and texturing methods with user specified parameters.

| 
|
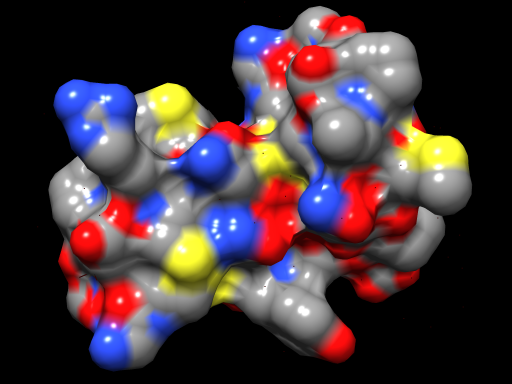
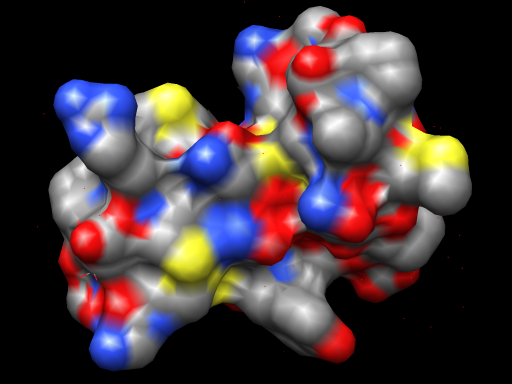
| Per-pixel lighting with an OpenGL fragment shader. | Normal OpenGL lighting without shaders. |
| 
|
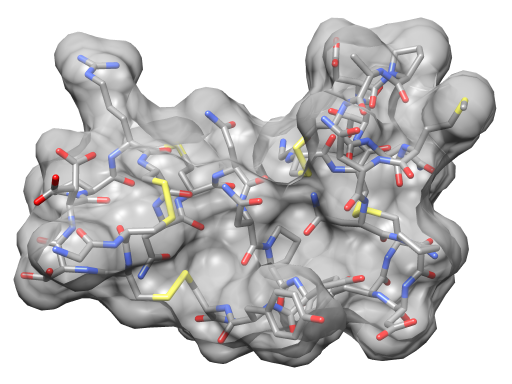
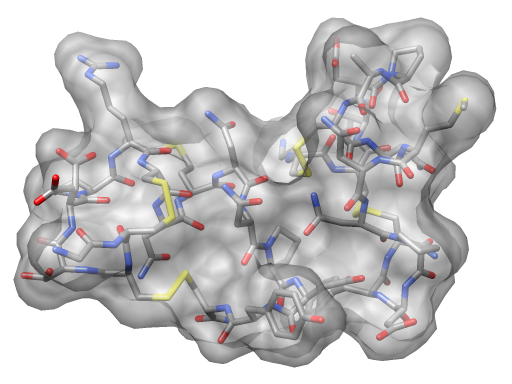
| Angle-dependent transparency with an OpenGL fragment shader. | Normal OpenGL transparency without shaders. |
Above Chimera shader images made with the following OpenGL fragment shader.
varying vec3 N;
varying vec3 v;
void main (void)
{
const int kl = 1; // Chimera key light is 1, fill light is 0.
const int fl = 0;
vec3 N1 = normalize(N); // Surface normal
vec3 L = normalize(gl_LightSource[kl].position.xyz); // Key light direction
vec3 Lf = normalize(gl_LightSource[fl].position.xyz); // Fill light direction
vec3 E = normalize(-v); // Eye direction
vec3 R = normalize(-reflect(L,N1)); // Reflection direction
// Ambient light
vec4 Iamb = gl_FrontLightProduct[kl].ambient; // Default light ambient = 0
// Diffuse light
vec4 Idiff = gl_Color * (gl_LightSource[kl].diffuse * max(dot(N1,L), 0.0)
+ gl_LightSource[fl].diffuse * max(dot(N1,Lf), 0.0));
// Specular light
vec4 Ispec = gl_FrontLightProduct[kl].specular
* pow(max(dot(R,E),0.0),0.3*gl_FrontMaterial.shininess);
// Scene light
vec4 Iscene = gl_Color * gl_LightModel.ambient; // Acm * Acs
// Angle dependent transarency
float a = 1.0 - pow(max(1.0-gl_Color.a,0.0), 1.0/max(abs(N1.z),0.01));
// Total color
gl_FragColor = vec4(Iscene.rgb + Iamb.rgb + Idiff.rgb + Ispec.rgb, a);
}
Acronym: GPGPU = General Purpose computing on GPU

For comparison, my older laptop graphics Radeon X1600 (Dec 2005) has 5 vertex and 12 fragment shaders, 10 GB/sec memory bandwidth, 128 bit bus width.
Comparison of graphics processing units (Wikipedia): Nvidia, ATI
Cost: GeForce 8800 GTX ~$500, Quadro FX 5600 ~$2500, Radeon HD 4870 ~$250.
| Model | Year | Code name | Fab (nm) | Transistors (Million) | Die Size (mm2) | Bus interface | Memory min (MiB) | Config core1 | Reference clock rate | Fillrate | Memory | Graphics library support (version) | GFLOPs (MADD/MUL) | TDP (Watts) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Core (MHz) | Shader (MHz) | Memory (MHz) | Pixel (MP/s) | Texture (MT/s) | Bandwidth reference (GB/s) | Bus type | Bus width (bit) | DirectX | OpenGL | |||||||||||
| GeForce 8800 GTX [7] | Nov 2006 | G80 | 90 | 681 | 484 | PCIe x16 | 768 | 128:642:24 | 575 | 1350 | 1800 | 13800 | 36800 | 86.4 | GDDR3 | 384 | 10 | 2.1 | 518 | 155 |
| Model | Code name | Fab (nm) | Bus interface | Memory max (MiB) | Core clock max (MHz) | Memory clock max (MHz) | Config core123 | Fillrate max (MT/s) | Memory | Graphics library support (version) | Features | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bandwidth max (GB/s) | Bus type | Bus width (bit) | DirectX | OpenGL | ||||||||||
| Quadro FX 56002 | G80 | 55 | PCIe 2.0 x16 | 1536 | 600 | 1600 | 196:32:24 | 38400 | 76.8 | GDDR3 | 384 | 10 | 2.0 | Stereo display, SLI, Genlock |
| Model | Year | Code name | Fab (nm) | Bus interface | Memory max (MiB) | Reference clock rate | Config core1 | Fillrate | Memory | Graphics library support (version) | Notes | GFLOPS3 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Core (MHz) | Memory (MHz) | Texture (GT/s) | Pixel (GP/s) | Bandwidth (GB/s) | Bus type | Bus width (bit) | DirectX | OpenGL | |||||||||
| Radeon HD 4870 | Jun 25, 2008[23] | RV770 XT | 55 | PCIe 2.0 x16 | 512,1024 | 750 | 9002 | 800(160x5):40:16 | 31.6[20] | 12.7 | 115.2[20] | GDDR52 | 256 | 10.1 | 2.1 | UVD2, PowerPlay | 1200[20] |
| Model | Year | Model Number | Code name | Fab (nm) | Bus interface | Memory max (MiB) | Core clock max (MHz) | Memory clock max (MHz) | Config core1 | Fillrate max (MT/s) | Memory | Graphics library support (version) | Notes | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bandwidth max (GB/s) | Bus type | Bus width (bit) | DirectX | OpenGL | ||||||||||||
| Mobility Radeon X1600 | Dec 2005 | M56 | RV530 | 90 | PCIe x16 | 256 | 445 | 350 | 5:12:4:4 | 1780 | 11.20 | GDDR3 | 128 | 9.0c | 2.0 | |
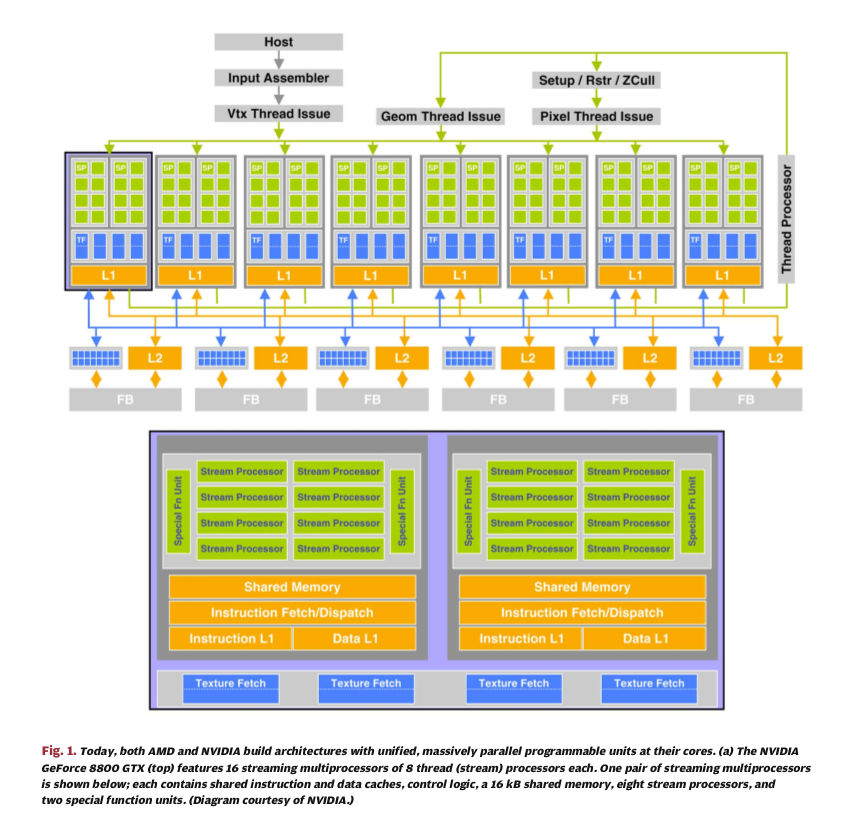
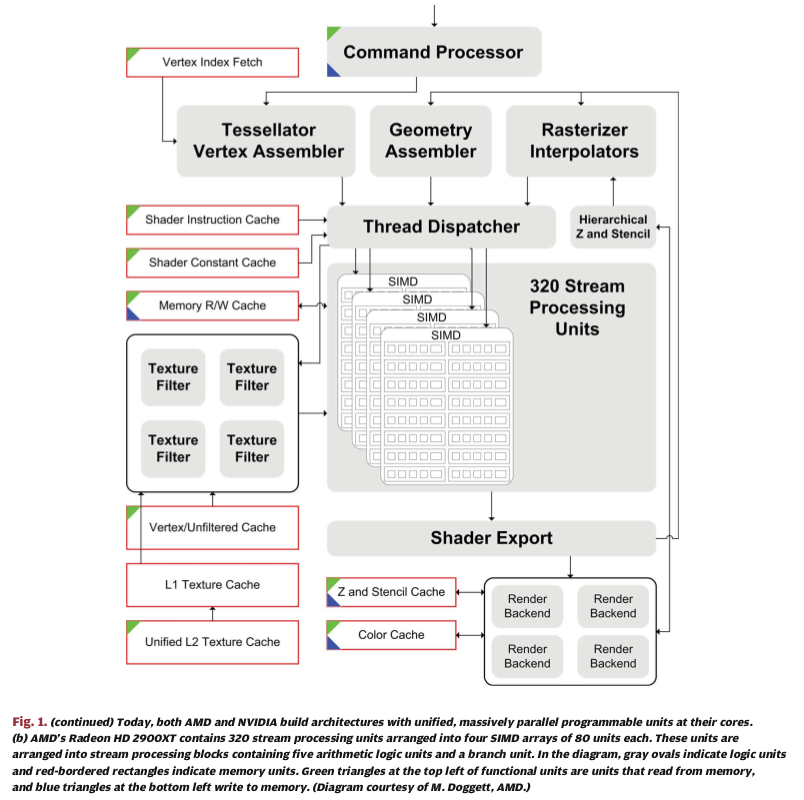
Above figures from:
GPU Computing: Graphics Processing Units -- powerful, programmable, and highly parallel -- are increasingly targeting general-purpose computing applications.
John D. Owens , Mike Houston , David Luebke , Simon Green, John E . Stone , and James C. Phillips
Proceedings of the IEEE, Vol. 96, No. 5, May 2008 p 879-899
Some explanations of stream processing, the GPU programming model, from Wikipedia.
Given a set of data (a stream), a series of operations (kernel functions) are applied to each element in the stream. Uniform streaming, where one kernel function is applied to all elements in the stream, is typical.
Stream processing allows parallel processing without explicitly managing allocation, synchronization, or communication among processing units.
Stream processing is especially suitable for applications that exhibit three application characteristics:
Some limitations mentioned in GPGPU Wikipedia article.
Modest performance gains. While GPGPU can achieve a 100-250x speedup vs a single CPU, only embarrassingly parallel applications will see this kind of benefit. Quoted speed-ups in published applications tend to be 5x - 20x.
Note: Chimera speed-up of Quadro FX 5600 (196 shader units) over Mobile Radeon X1600 (17 shader units) is just 2x.
Slow or no double precision. Current top-end GPUs from Nvidia and AMD emphasize single-precision (32-bit) computation; double-precision (64-bit) computation executes much slower. Also 32-bit and 64-bit operations are sometimes (often?) not IEEE compliant.
Limited branching and looping. In regular programs it is possible to control the flow of the program using if-then-else statements and various forms of loops. Such flow control structures have only recently been added to GPUs. Recent GPUs allow branching, but usually with a performance penalty. The branching is done by splitting a group of streams executing the same instructions into two groups according to which branch is taken by each stream processor. A dedicated "branching unit" manages the split.
Difficult debugging. Can't print from the GPU program. Instead computed results as images.
Graphics card and operating-specific code. Nvidia and AMD offer competing GPU programming languages that only work with their products.
Most programming languages for stream processors start with C or C++ and add extensions which provide specific instructions to allow application developers to tag kernels and/or streams.
CUDA (Compute Unified Device Architecture) Nvidia GPU programming language.
Stream SDK AMD GPU programming languague (earlier sdk called CTM).
OpenCL 1.0 (Dec 8, 2008 spec). Apple parallel computing language for GPUs. To be introduced in Mac OS 10.6. AMD (ATI) and Nvidia are adding full support (Dec 2008).
Most shading languages for graphics GPU programs are similarly based on C.
Cg (Nvidia) [MGAK03], HLSL (Microsoft) [Mic05a], and the OpenGL Shading Language [KBR04] all abstract the capabilities of the underlying GPU and allow the programmer to write GPU programs in a more familiar C-like programming language. They do not stray far from their origins as languages designed to shade polygons. All retain graphics-specific constructs: vertices, fragments, textures, etc. Cg and HLSL provide abstractions that are very close to the hardware, with instruction sets that expand as the underlying hardware capabilities expand. The OpenGL Shading Language was designed looking a bit further out, with many language features (e.g. integers) that do not directly map to hardware available today. (Owens JD, 2008).
www.gpgpu.org. "The goal of this page is to catalog the current and historical use of GPUs for general-purpose computation."
Articles
* GPGPU (369)
o Advanced Rendering (37)
+ Global Illumination (13)
+ Image-Based Modeling & Rendering (5)
o Audio and Signal Processing (4)
o Computational Geometry (14)
+ GIS (1)
+ Surfaces and Modeling (4)
o Computer Architecture (2)
o Conferences (32)
o Contests (2)
o Data Parallel Algorithms (7)
o Database (7)
+ Sort & Search (2)
o GPUs (10)
o High-Level Languages (27)
o Image And Volume Processing (43)
+ Compression (1)
+ Computer Vision (10)
o Med & Bio (2)
o Miscellaneous (44)
+ Books (6)
+ Courses (15)
+ Developer Resources (14)
+ Journals (2)
+ Research Groups (3)
o Pattern Matching (1)
o Press (13)
o Scientific Computing (84)
+ Data Compression (2)
# Data Structures (1)
+ Dynamics Simulation (4)
+ Mathematics (1)
+ Numerical Algorithms (8)
o Site News (8)
o Stream Processing (1)
o Tools (26)
First generation GPU core September 2006.
Second generation GPU core April 2008.
ATI X1600 - X1900 GPUs only.
Windows XP and Vista only.
What about video cards with other (non-ATI) chipsets?
The R580 (in the X1900XT, etc.) performs particularly well for molecular dynamics, due to its 48 pixel shaders. Currently, other cards (such as those from nVidia and other ATI cards) do not perform well enough for our calculations as they have fewer pixel shaders. Also, nVidia cards in general have some technical limitations beyond the number of pixel shaders that makes them perform poorly in our calculations.
Is the GPU client for Windows XP only? Has it been tested on other OS's like Linux, Mac, and Vista?
We will launch with Windows XP (32 bit only) support due to driver and compiler support issues. In time, we hope to support Linux as well. Macintosh OSX support is much further out, as the compilers and drivers we need are not supported in OSX, and thus we cannot port our code until that has been resolved. Users have reported the GPU client works in Vista, but due to a different DX version, the performance characteristics vary slightly from Windows XP.
Most GPU articles are on medical imaging. I include only a few here.
Most articles are published in IEEE Transactions on Visualization and Computer Graphics.
Many GPU medical imaging applications from annual conference of the Medical Image Computing and Computer Assisted Intervention Society. Also many from a conference series Medicine Meets Virtual Reality.
On modelling of anisotropic viscoelasticity for soft tissue simulation: Numerical solution and GPU execution.
Taylor ZA, Comas O, Cheng M, Passenger J, Hawkes DJ, Atkinson D, Ourselin S.
Med Image Anal. 2008 Oct 17. [Epub ahead of print]
Finite element modeling for biomechanical image registration
and interactive surgical simulation.
Automatic labeling of anatomical structures in MR FastView images using a statistical atlas.
Fenchel M, Thesen S, Schilling A.
Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2008;11(Pt 1):576-84.
Deformable matching of medical imaging data to atlas of anatomical structures.
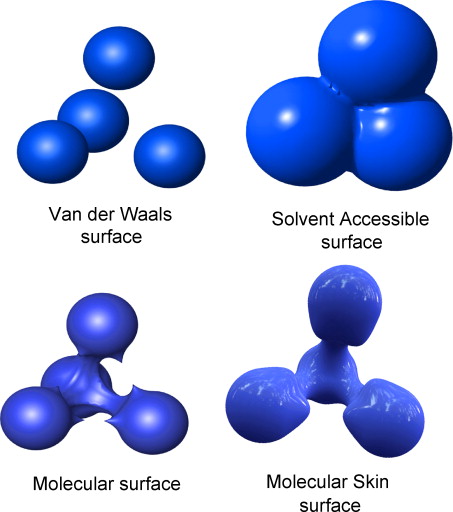
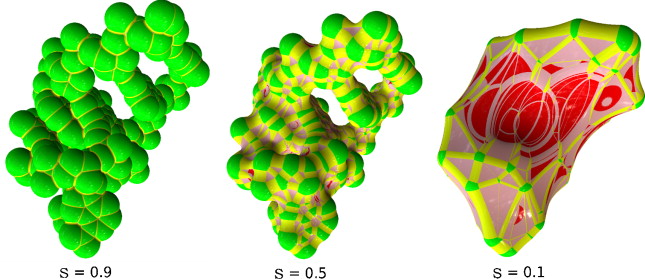
MetaMol: high-quality visualization of molecular skin surface.
Chavent M, Levy B, Maigret B.
J Mol Graph Model. 2008 Sep;27(2):209-16. Epub 2008 Apr 29.
Raycasting computation of molecular skin surface, a smoother variant of Connolly surface, from analytic quartic surface. Eliminates two levels of sampling
when rendering with a triangulation.
Performance evaluation of image processing algorithms on the GPU.
Castaño-Díez D, Moser D, Schoenegger A, Pruggnaller S, Frangakis AS.
J Struct Biol. 2008 Oct;164(1):153-60. Epub 2008 Jul 24.
GPU implementations of common algorithms used for three-dimensional
image processing: spatial transformations, real-space and Fourier
operations, as well as pattern recognition procedures, reconstruction
algorithms and classification procedures. 10-20x speed-up.
A streaming narrow-band algorithm: interactive computation and visualization of level sets.
Lefohn AE, Kniss JM, Hansen CD, Whitaker RT.
IEEE Trans Vis Comput Graph. 2004 Jul-Aug;10(4):422-33.
Level-set methods use partial differential equations to deform isosurfaces
-- used of volume data segmentation. Requires fast interactive computation
to hand-tune parameters.
CUDA compatible GPU cards as efficient hardware accelerators for Smith-Waterman sequence alignment.
Manavski SA, Valle G.
BMC Bioinformatics. 2008 Mar 26;9 Suppl 2:S10.
Smith-Waterman alignment takes time proportional to product of lengths of
sequences -- much slower than BLAST and FASTA. Tried Smith-Waterman on a
machine with two Geforce 8800 GTX cards. Equaled BLAST on length 500 sequences.
Two-electron integral evaluation on the graphics processor unit.
Yasuda K.
J Comput Chem. 2008 Feb;29(3):334-42.
Evaluate the Coulomb potential in the ab initio density functional calculation.
Evaluates limits of single precision floating point. Uses Geforce 8800 GTX to
find energies of Taxol and Valinomycin.
High-throughput sequence alignment using Graphics Processing Units.
Schatz MC, Trapnell C, Delcher AL, Varshney A.
BMC Bioinformatics. 2007 Dec 10;8:474.
GPU implementation
of MUMmer - a program for
rapidly aligning entire genomes. Can find all 20-basepair or longer
exact matches between a pair of 5-megabase genomes. Handle
1000s of contigs from a shotgun sequencing project.
Accelerating molecular modeling applications with graphics processors.
Stone JE, Phillips JC, Freddolino PL, Hardy DJ, Trabuco LG, Schulten K.
J Comput Chem. 2007 Dec;28(16):2618-40.
Calculation of long-range electrostatics and nonbonded forces for
molecular dynamics simulations. Coulomb-based ion placement and
calculation of time-averaged potentials from molecular dynamics
trajectories.
Two-level approach to efficient visualization of protein dynamics.
Daae Lampe O, Viola I, Reuter N, Hauser H.
IEEE Trans Vis Comput Graph. 2007 Nov-Dec;13(6):1616-23.
GPU computation of atom positions based on rotation of residues.
Multi-level graph layout on the GPU.
Frishman Y, Tal A.
IEEE Trans Vis Comput Graph. 2007 Nov-Dec;13(6):1310-9.
Force-directed graph layout.
Real-time 3D computed tomographic reconstruction using commodity graphics hardware.
Xu F, Mueller K.
Phys Med Biol. 2007 Jun 21;52(12):3405-19. Epub 2007 May 17.
Back-projection calculation for medical imaging. Same technique as single-particle EM.
Fast collision detection based on nose augmentation virtual surgery.
Xie K, Yang J, Zhu YM.
Comput Methods Programs Biomed. 2007 Oct;88(1):1-7. Epub 2007 Aug 13.
"Collision detection is the key technology in nose augmentation surgery simulation system, which can avoid incorrect intersection between bones and the implant model."
GPU-friendly marching cubes for visualizing translucent isosurfaces.
Xie Y, Heng PA, Wang G, Wong TT.
Stud Health Technol Inform. 2007;125:500-2.
Computes and draws isosurface directly on the graphics card. Allows
multiple transparent surfaces. No depth sorting needed since contours
are found layer by layer back to front.
Implementation and performance evaluation of reconstruction algorithms on graphics processors.
Castaño Díez D, Mueller H, Frangakis AS.
J Struct Biol. 2007 Jan;157(1):288-95. Epub 2006 Sep 1.
Electron tomography and single-particle EM recontruction on the GPU.
Tries 3 algortithms, compared speeds to CPU.
Failure of a traffic control "fatality" sign to affect pedestrians' and motorists' behavior.
Harrell WA, David-Evans M, Gartrell J.
Psychol Rep. 2004 Dec;95(3 Pt 1):757-60.
Word "gpu" is in authors email address.
A Survey of General-Purpose Computation on Graphics Hardware.
John D. Owens, David Luebke, Naga Govindaraju, Mark Harris, Jens Krüger, Aaron E. Lefohn, and Tim Purcell.
Computer Graphics Forum, volume 26, number 1, 2007, pp. 80-113.
Speeding Up Molecular Docking Calculations Using Consumer Graphics Hardware. (Efficient Ant Colony Optimization Algorithms for Structure- and Ligand-Based Drug Design. Oliver Korb PhD thesis, University of Konstanz, 2008) Posted: 18 Nov 2008.
GPU acceleration of cutoff pair potentials for molecular modeling applications. (C. I. Rodrigues, D. J. Hardy, J. E. Stone, K. Schulten, W. W. Hwu., GPU acceleration of cutoff pair potentials for molecular modeling applications. Proceedings of the 2008 Conference On Computing Frontiers, pp.273-282, 2008.) (http://www.ks.uiuc.edu/Research/gpu/). Posted: 25 May 2008
CUDA compatible GPU cards as efficient hardware accelerators for Smith-Waterman sequence alignment. (Svetlin A. Manavski, Giorgio Valle, CUDA compatible GPU cards as efficient hardware accelerators for Smith-Waterman sequence alignment, BMC Bioinformatics 2008, 9(Suppl 2):S10 (26 March 2008)).
Quantum Chemistry on GPUs. (Quantum Chemistry on Graphical Processing Units. 1. Strategies for Two-Electron Integral Evaluation, Ivan S. Ufimtsev and Todd J. Martínez, J. Chem. Theory Comput., 4 (2), 222 -231, 2008. doi:10.1021/ct700268q). Posted: 01 Apr 2008.
Accelerating Resolution-of-the-Identity Second-Order Møller-Plesset Quantum Chemistry Calculations with Graphical Processing Units. (Accelerating Resolution-of-the-Identity Second-Order Møller-Plesset Quantum Chemistry Calculations with Graphical Processing Units. Vogt, L., Olivares-Amaya, R., Kermes, S., Shao, Y., Amador-Bedolla, C., and Aspuru-Guzik, A. J. Phys. Chem. A, 2008, DOI: 10.1021/jp0776762). Posted: 10 Feb 2008
High-throughput sequence alignment using Graphics Processing Units. (High-throughput sequence alignment using Graphics Processing Units, Schatz, M.C., Trapnell, C., Delcher, A.L., Varshney, A. (2007), BMC Bioinformatics 8:474.). Posted: 10 Feb 2008
Genome Technology Article about GPGPU: "Not Just for Kids Anymore". This article at Genome Technology gives a brief overview of GPGPU, with a focus on biological information processing using NVIDIA CUDA Technology. The article discusses the results from UIUC's NAMD / VMD project and neurological simulation company Evolved Machines. Posted: 10 Sep 2007
Accelerating molecular modeling applications with graphics processors. ( Accelerating molecular modeling applications with graphics processors , John E. Stone, James C. Phillips, Peter L. Freddolino, David J. Hardy, Leonardo G. Trabuco, and Klaus Schulten. Journal of Computational Chemistry (In press)) Posted: 10 Aug 2007
What are the practical limitations of using GPU computation in production software, as opposed to use in research prototypes?
I didn't read anything that directly talked about this.
GPU computation is feasible in some production applications such as computer games and video processing.
For scientific applications, I think only highly compute bound applications which take days, weeks or months of CPU time are good candidates for GPGPU.
GPU computation for graphics (shaders) are ready for production use now in any OpenGL applications.