This tutorial describes generating high-quality images of a protein surface colored to show hydrophobicity and electrostatic potential. Note there are several routes to the same or similar results. See also: presets, tips on preparing images
Start Chimera and show the Command Line (for example, with Favorites... Command Line). Fetch Protein Data Bank entry 3eeb:
Command: open 3eebThe structure is a toxin that is activated by binding inositol hexakisphosphate (IHP), a highly negatively charged small molecule. As might be expected, the binding site is polar and lined with many positively charged groups. This can be illustrated by coloring the surface to show its properties.
Move and scale the structure as you wish throughout the tutorial. Delete one of the copies of the structure, chain A, and solvent:
Command: delete :.aResize the window as desired, either by dragging its lower right corner with the mouse or by using the command windowsize. The window dimensions define the aspect ratio (width:height) of output images, but image resolution (pixel dimensions) can be specified independently when an image is saved.
Command: del solvent
 |
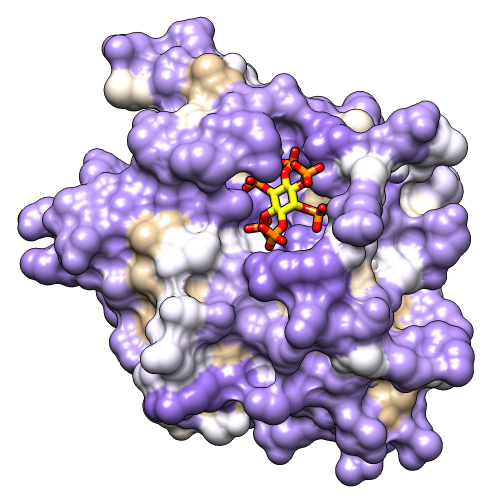
Use the “hydrophobicity surface” preset to display the molecular surface colored by amino acid hydrophobicity:
Command: preset apply int 3Colors are mapped to Kyte-Doolittle hydrophobicity: from dodger blue for the most hydrophilic, to white, to orange red for the most hydrophobic. The IHP molecule looks somewhat like a distorted snowflake. The IHP-binding pocket is primarily blue, indicating its hydrophilic character. However, as shown in the figure, we will use different colors than in the preset.
A nearby sodium ion and the heteroatoms in IHP are colored by element. Hide the ion, make the sticks fatter, and make ligand carbons yellow:
Command: ~disp ionsDifferent colors can be mapped to this kdHydrophobicity attribute of residues using the command rangecolor:
Command: setattr m stickScale 2
Command: color yellow ligand & C
Command: rangecol kdHydrophobicity min medium purple 0 white max tanNow the most polar residues are medium purple and the most hydrophobic are tan. (The same thing can be done in the Render by Attribute graphical interface; for a similar process, see the B-Factor Coloring image tutorial.)
The built-in colors can be viewed here or by choosing Actions... Color... all options from the menu and in the resulting dialog, checking the option to Show all colors. Additional colors can be created with the command colordef.
Use a publication preset for nice image settings, including white background, black outlines, and increased smoothness and shininess:
Command: preset apply pub 1The example image was saved from a 500x500-pixel window using File... Save Image with default settings.
 |
 |
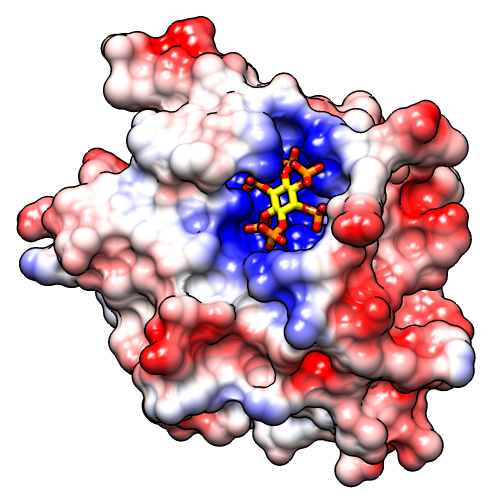
If not already done, show the molecular surface and use a publication preset:
Command: surfStart Coulombic Surface Coloring (under Tools... Surface/Binding Analysis), choose the surface model, and click OK.
Command: preset apply pub 1
The publication presets provide a nice starting point, but you might want to adjust some settings, such as the thickness of the black outlines:
Command: set silhouette_width 3If you prefer a simple “line drawing” appearance, try ambient lighting:
Command: light mode ambientTo restore default lighting:
Command: light restore Chimera defaultThe example images were saved from a 500x500-pixel window using File... Save Image with default settings.
There is a limit to how thick lines such as silhouette edges, wire, and mesh can be drawn while rendering an image. If supersampling is done, the image is initially drawn at a larger size than requested (3x3 by default) and then sampled back down to the final size. In large and/or supersampled images, lines may be thinner than expected. The File... Save Image dialog reports the effective maximum linewidth. It may be possible to achieve the desired thickness by reducing the supersampling level or the pixel dimensions of the image.
Chimera does not perform Poisson-Boltzmann electrostatics calculations, but it can read a grid file or “map” of electrostatic potential values output by another program such as DelPhi, APBS or UHBD. Surfaces can be colored by the map values using Surface Color or the command scolor. An example map file (3eebB.phi, calculated with DelPhi for the same structure as in the tutorial above) is provided for those who wish to try this approach.