



assemblies
cryoEM maps
Tom Goddard
December 15, 2006

| 
| 
| 
|
| Proteins and sequences | Crystallographic maps | Molecular assemblies | Single particle cryoEM maps |

| 
| 
| 
|
| EM tomography cell components | EM tomography cells | Light microscopy | |
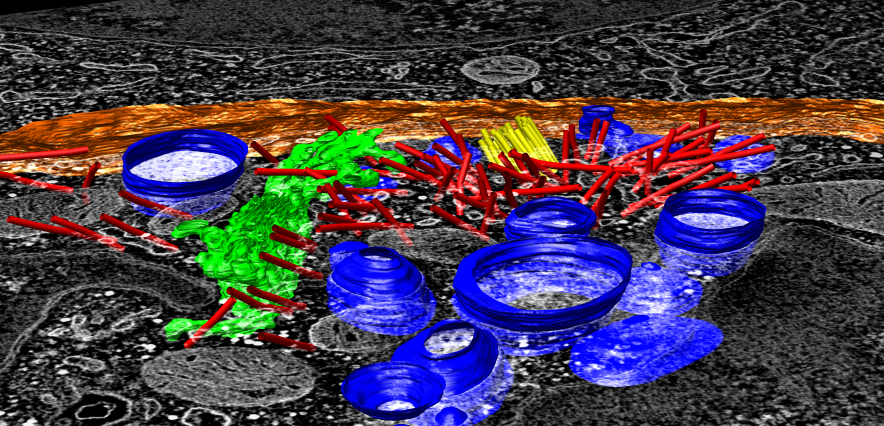
Chimera has map display and analysis capabilities developed for single particle cryoEM reconstructions, but none developed for tomography of cells. What does Chimera do better than Analyze, IMOD, or Amira?
Screenshots show Chimera tools developed by Wah Chiu's lab.

| 
|
Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM.
Centrosome polarization delivers secretory granules to the immunological synapse.
Nature. 2006 Sep 28;443(7110):462-5.
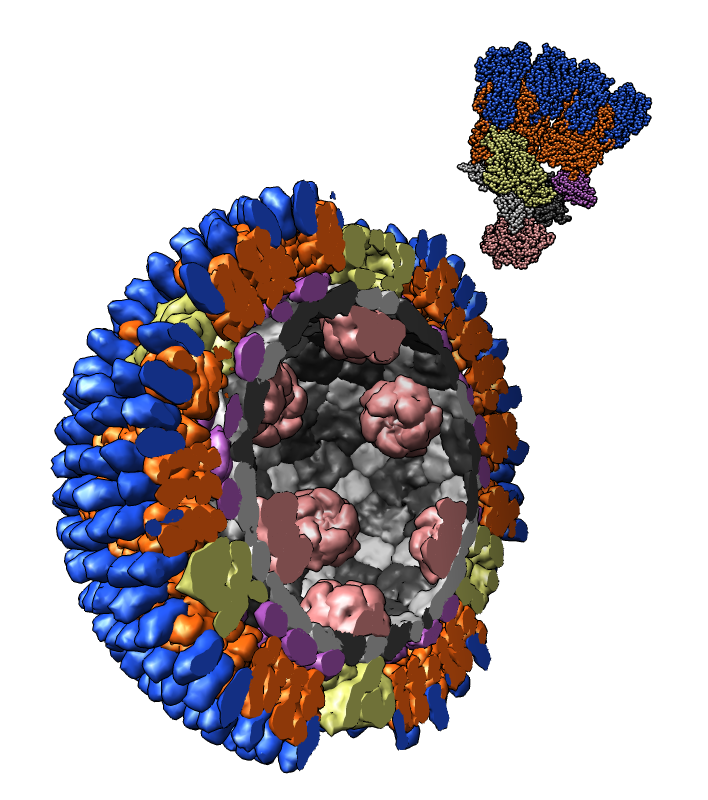
Atomic model of orthoreovirus (PDB 2cse) cut in half with one icosahedral asymmetric unit shown at upper right.
Rendering cartoon surfaces for 1620 proteins is interactive. Slow for 10,000 protein systems.
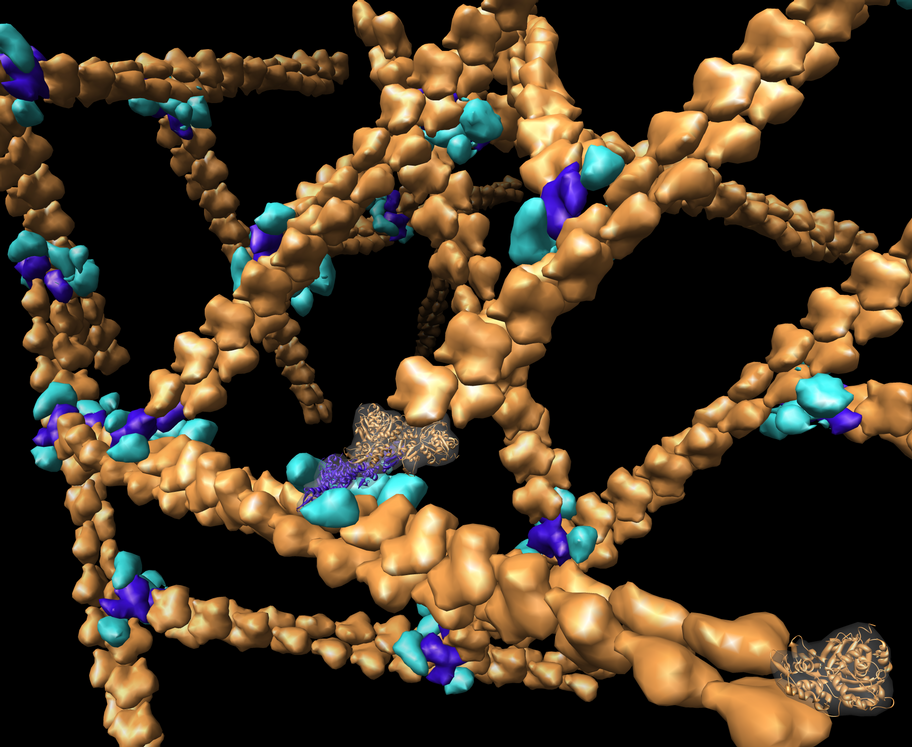
Branching actin filaments. Action monomers in orange. ARP2/3 branching complex in light and dark blue. (monomer PDB 1atn, ARP2/3 PDB 1k8k, filament PDB 1alm). Branching created by computational model, not from experimental data.
Rhinovirus with fragment of cellular receptor (blue) (PDB 1v9u). Exploring contacts between receptor and virus.

| 
|

| 
|
Measuring lengths of spines on neuron dendrite using hand-placed markers. IMOD segmentation of confocal microscopy data from Maryanne Martone. Forked and irregular spine shapes. Random spine colors to distinguish neighbors.

| 
| 
|
Time-series of dividing drosophila cells with fluorescently labeled chromosomes from Sedat lab. Each frame is 3 dimensonal. Wide field deconvolution microscopy. Animations: one cell, many cells.

| 
|

|
Tom Ferrin - lab director.
Greg Couch - molecular graphics.
Conrad Huang - project leader.
Elaine Meng - documentation.
Eric Pettersen - molecular sequence and structure.
Stephen Fuller (Oxford) - Immune synapse (CCDB 3632).
Ken Downing (Lawrence Berkeley Labs) - Microtubule map.
Maryanne Martone (UC San Diego) - Neuron dendrite.
Yuri Strukov and John Sedat (UC San Francisco) - Chromosomes.