
Tom Goddard
July 8, 2014
Ambient occlusion lighting darkens the crevices in molecules, maps and surfaces, greatly improving the 3-dimensional appearance. This is like depth cuing which dims the scene far from the viewer, but instead dims the scene in recesses where light would not reach. This will be a noteworthy new feature of ChimeraX, especially if it were enabled by default for some types of models. Here are example images testing techniques for calculating the lighting.
All the examples compute a low resolution density map for the model, then color the model darker in the high density regions. Another approach using shadowing along hundreds of directions should be tried.
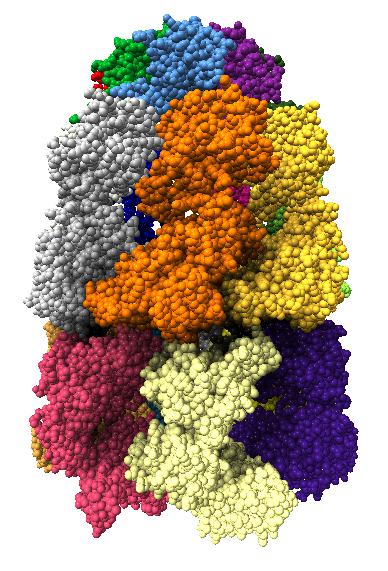
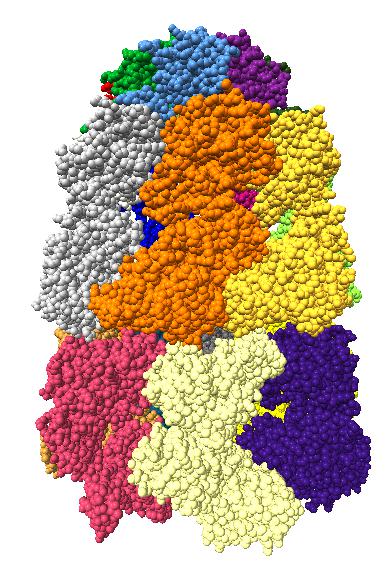
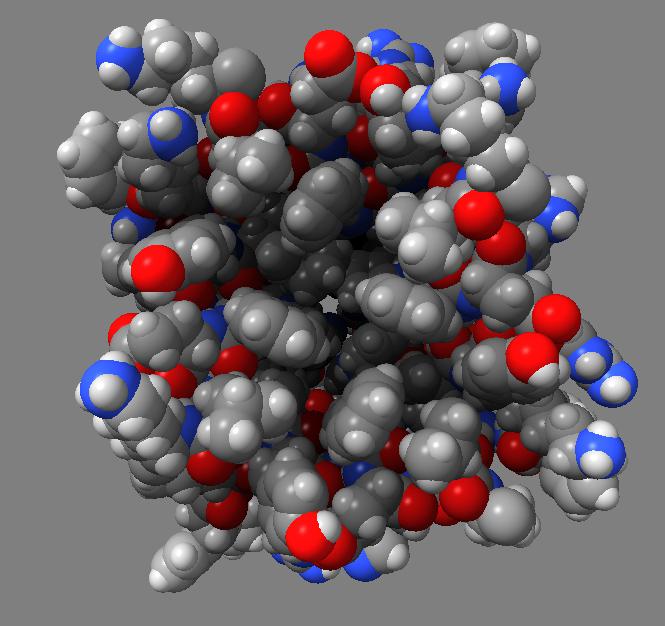
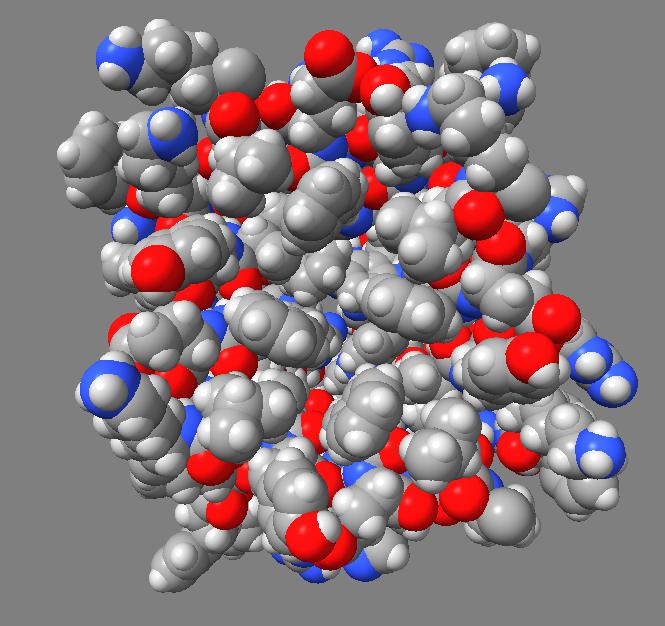
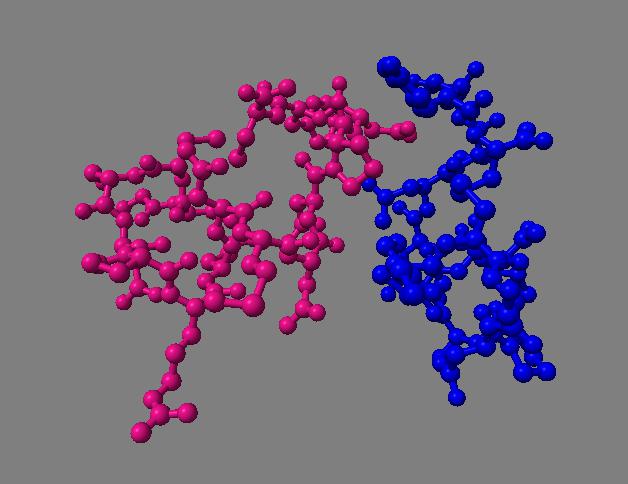
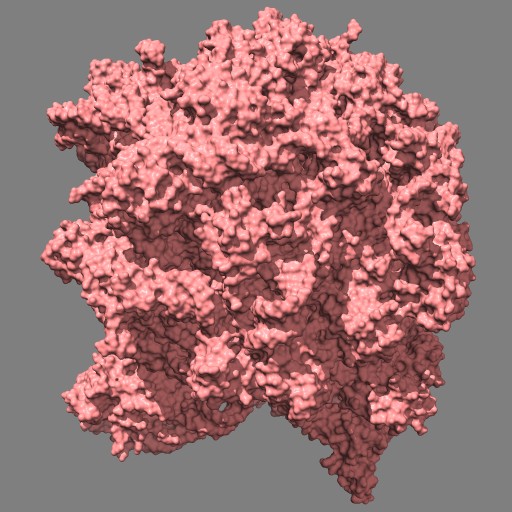
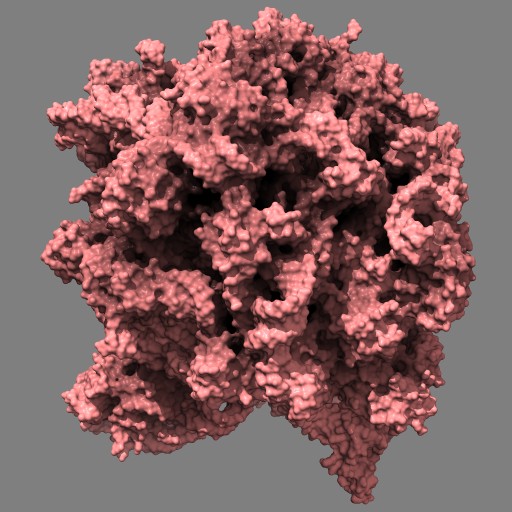
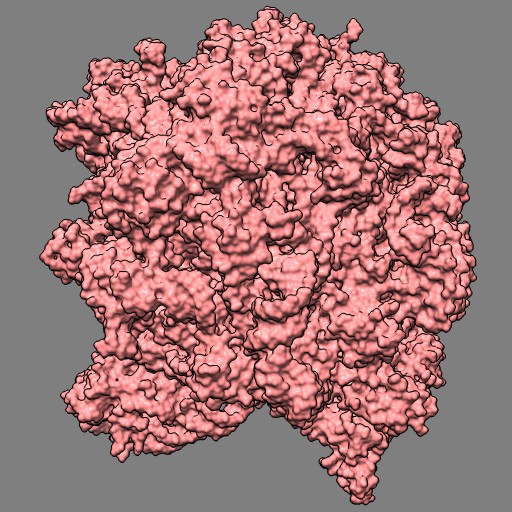
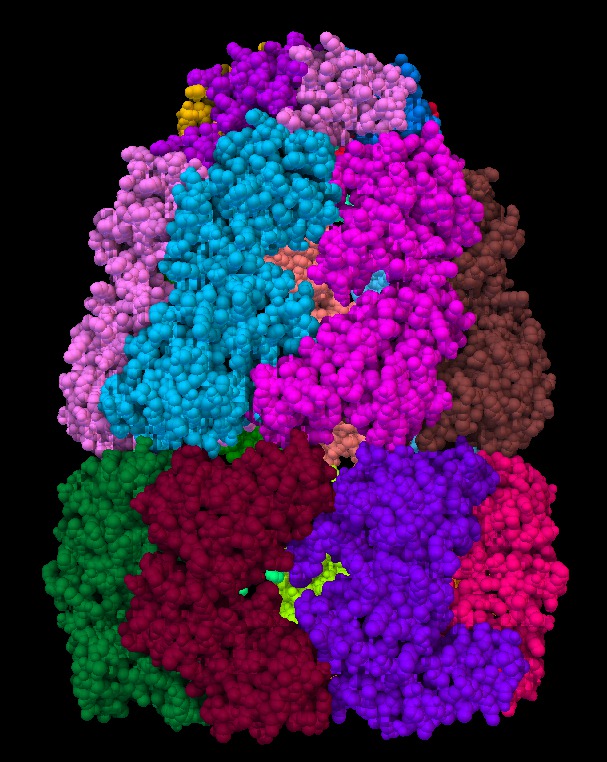
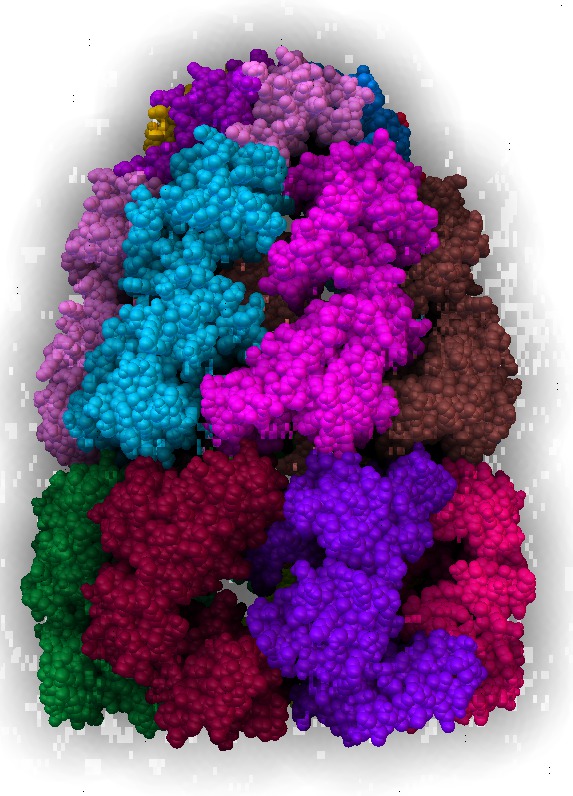
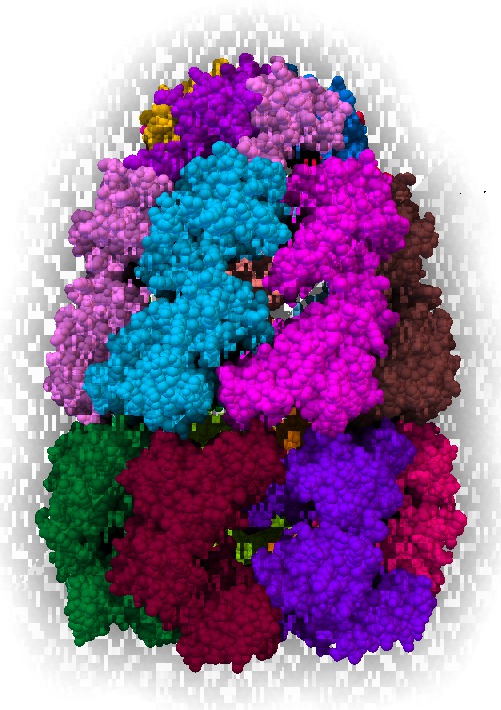
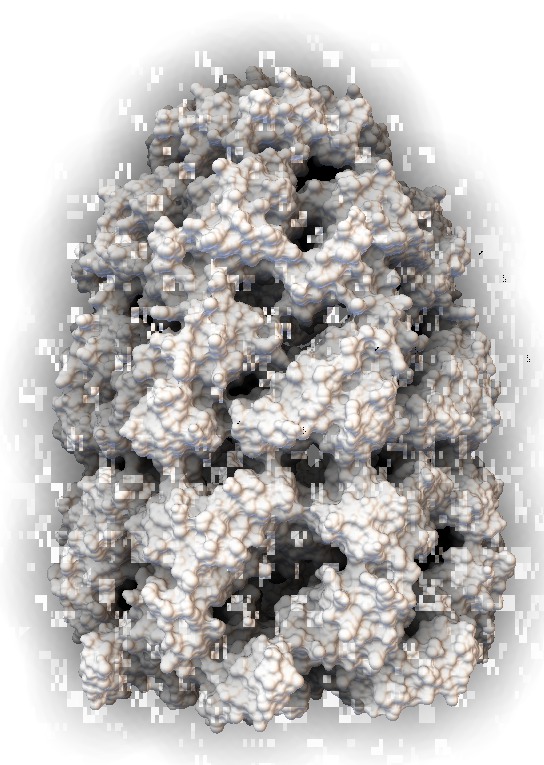
Comparison of ambient occlusion to normal Chimera lighting.
| Ambient occlusion | Normal lighting | Ambient occlusion | Normal lighting |
|---|---|---|---|

| 
| 
| 
|

| 
| 
| 
|

| 
| 
| 
|

| 
|
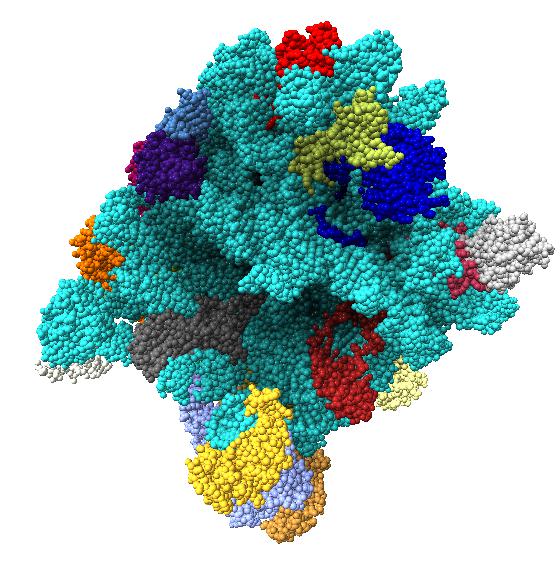
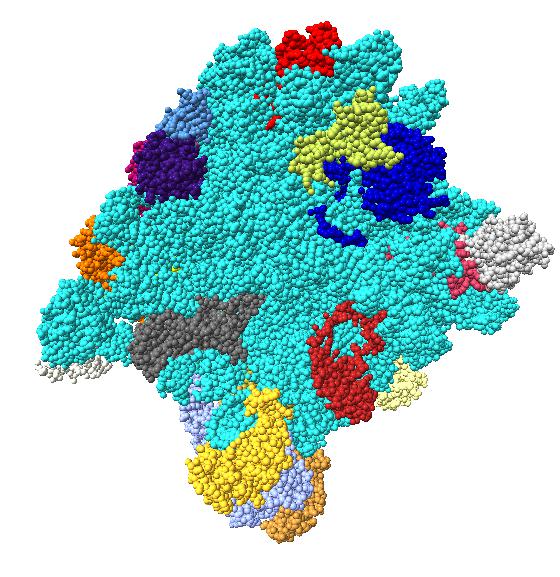
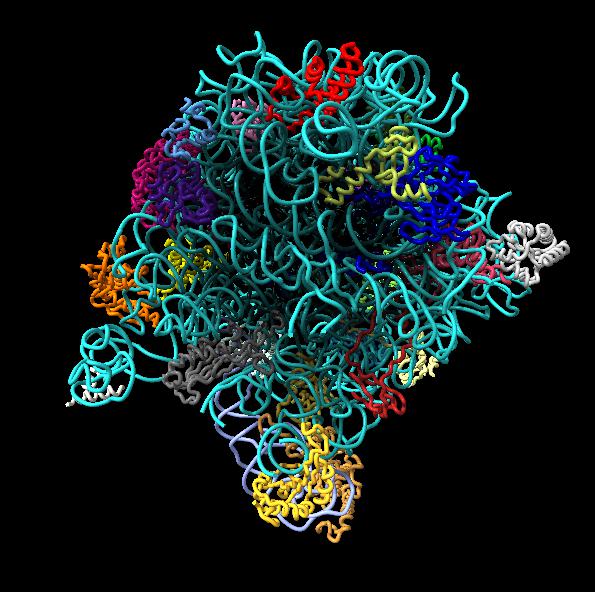
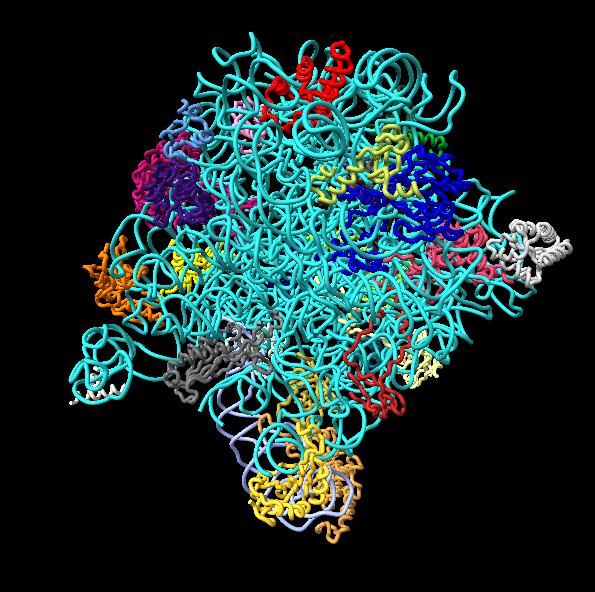
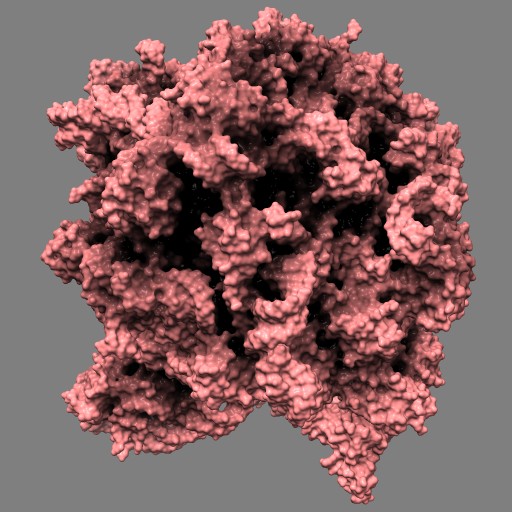
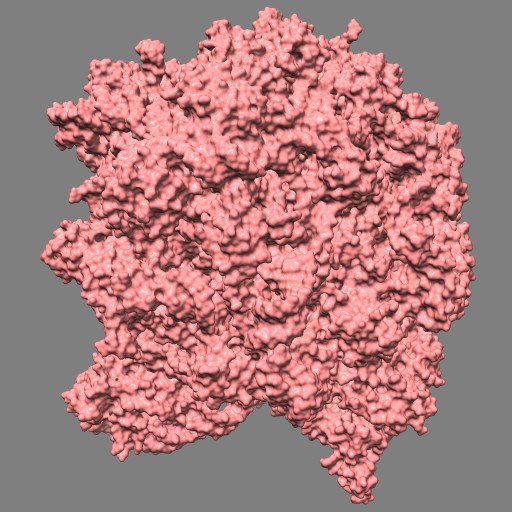
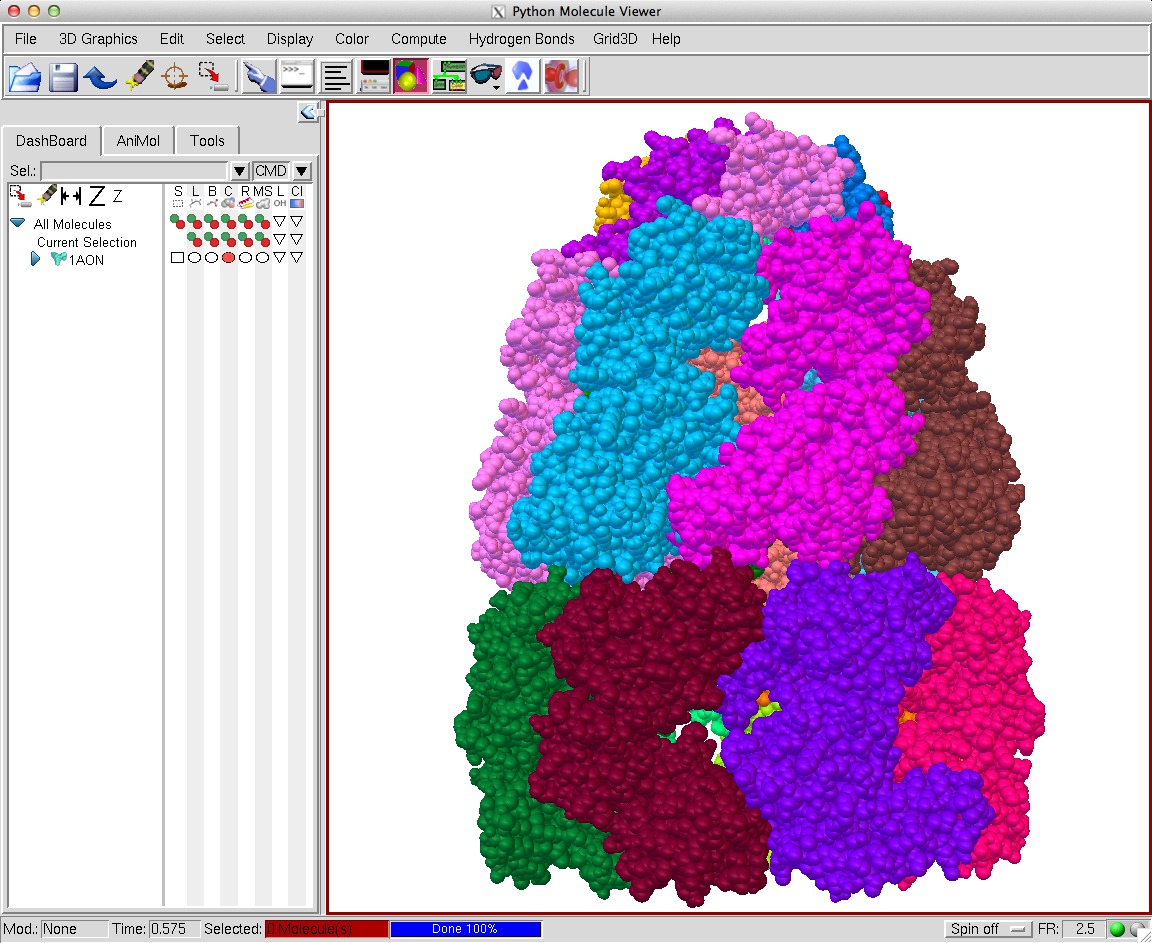
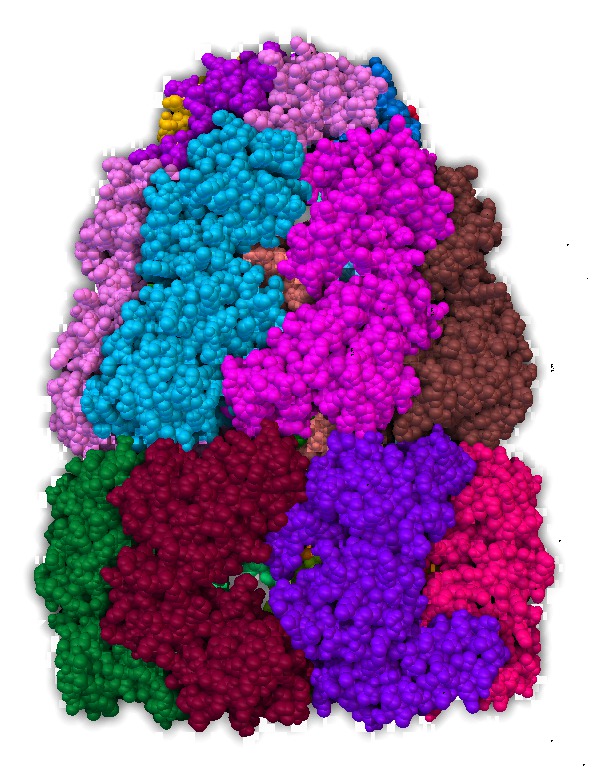
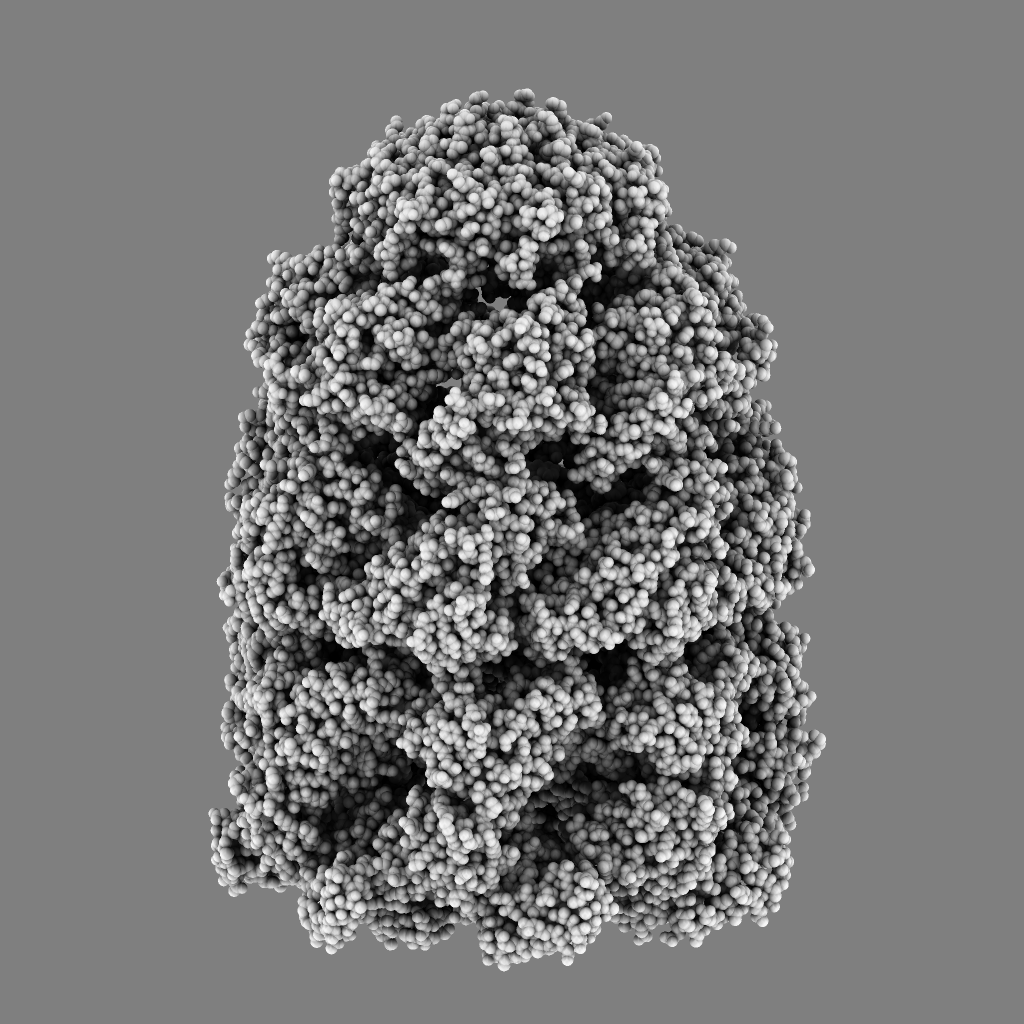
Images made in Hydra with command "ambient #1" except for chaperonin (upper left) and ribosome ribbon (middle right) which used "ambient #1 light 0.7". PDBs chaperonin 1aon, influenza ion channel 2kwx, ribosome 1jj2, conotoxin 1a0m, hiv capsid 3j3q. Computation of ambient occlusion is fast, 0.007 seconds for 100,000 atom ribosome structure, and 0.18 seconds for 2.4 million atom hiv capsid structure. Densities were computed by binning atoms using small map sizes, about 10 to 30 grid points wide.
| 
| 
| 
| 
|
| Ambient occlusion. Movie. | Normal lighting. Movie. | Shadows. Movie. | Ambient occlusion and shadows. Movie. | Silhouette edges. |
This example uses a gaussian smoothed ones mask of solvent excluded surfaces of a ribosome to color the surface. Made in Chimera.
open 1jj2 surf #0 grid .5 mask ones #1 spacing 10 vop gauss #2 sdev 10 scolor #1 volume #3 cmap 0.2,pink:0.5,black
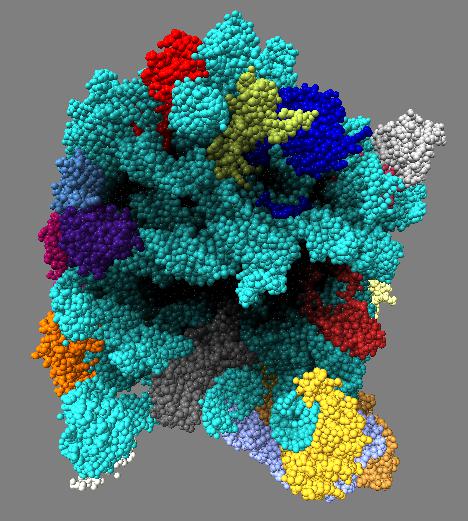
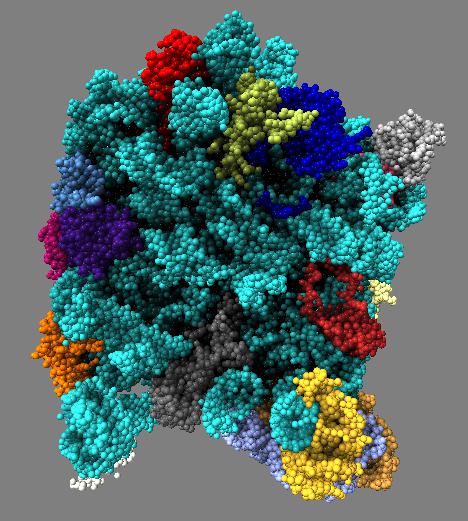
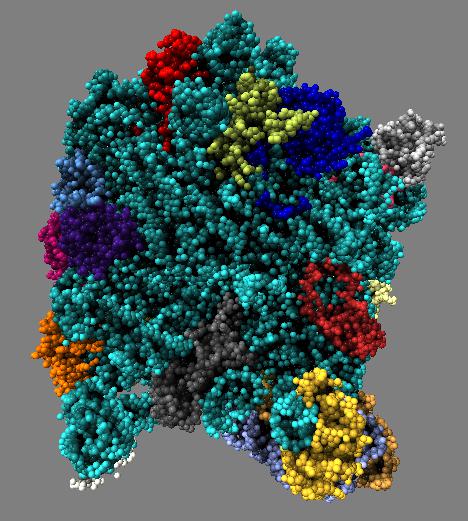
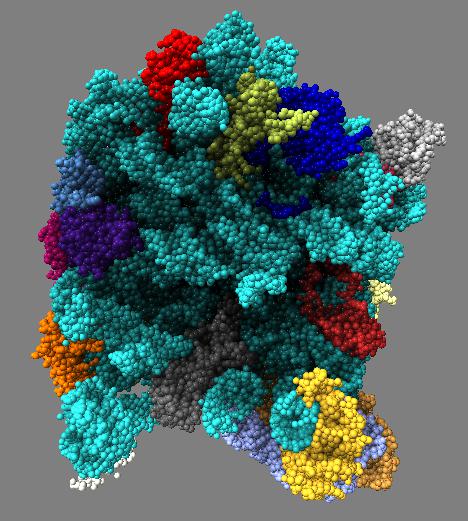
Comparison of ambient occlusion darkening recesses of different sizes. Fineness parameter is a fraction of the full width of the molecule and recesses of around that size are darkened.
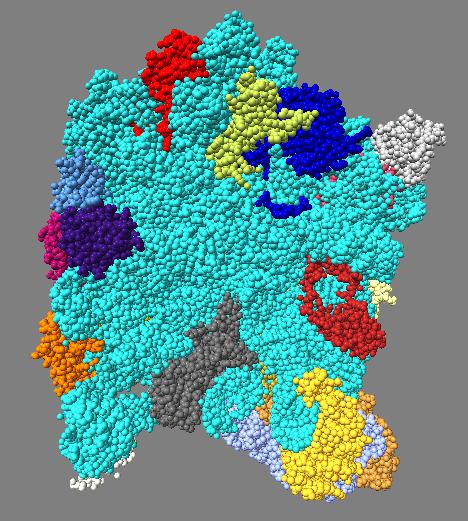
| 
| 
| 
| 
|
| Normal lighting. | Fineness 0.3 | Fineness 0.1 | Fineness 0.05 | Fineness 0.3 and 0.1 |
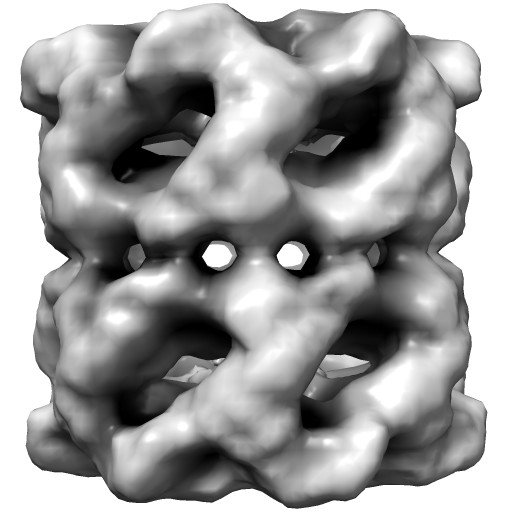
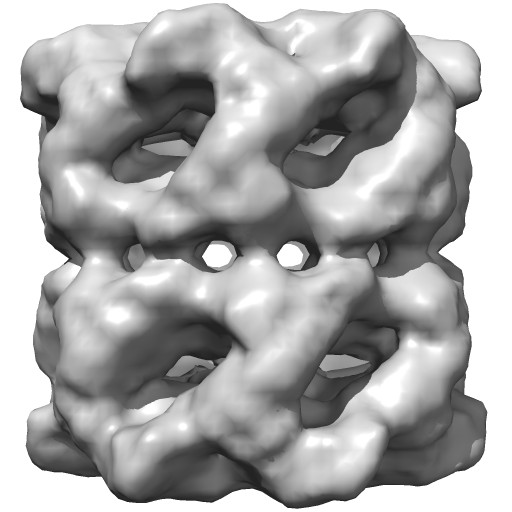
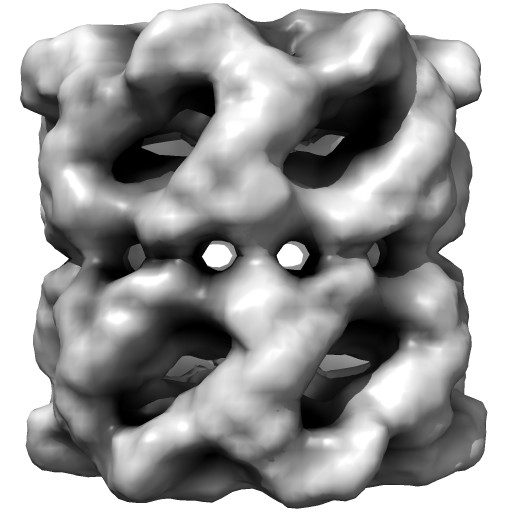
Images made in Hydra. Command "ambient #1 fineness 0.1 dark 0.2" except for combined 0.3 and 0.1 image that used "ambient #1 fine 0.3" followed by "ambient #1 fine 0.1".

| 
| 
| 
|
| Normal lighting. Movie. | Ambient occlusion. Movie. | Shadows. Movie. | Ambient occlusion and shadows. Movie. |
Groel images made with Chimera. "vop bin #0 bin 7" and "scolor #0 vol #1 cmap 0.5,gray:1,black". EMDB 1080.
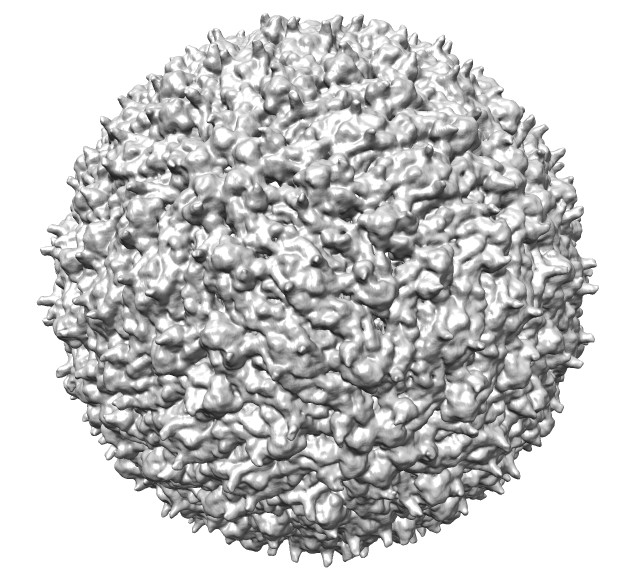
| 
| 
| 
|
| Normal lighting | Ambient occlusion | Shadows | Radially colored |
Dengue virus images made with Chimera. "vop bin #0 bin 7" and "scolor #0 vol #1 cmap 0.3,gray:0.4,black". Ambient occlusion essentially gives radial coloring for round virus. EMDB 5520.
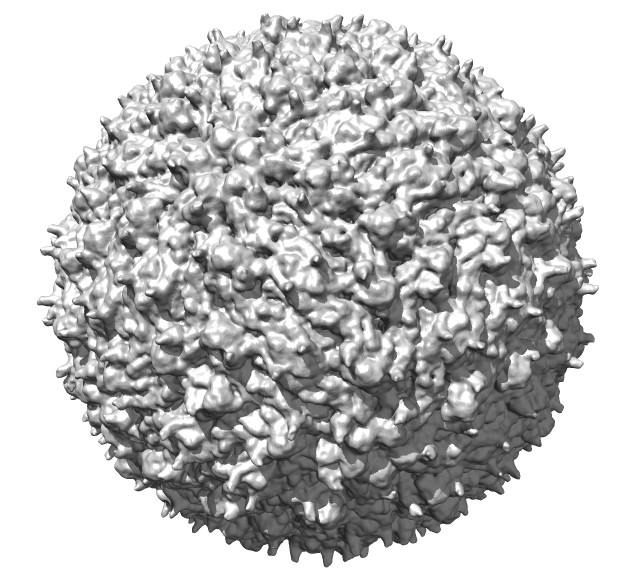
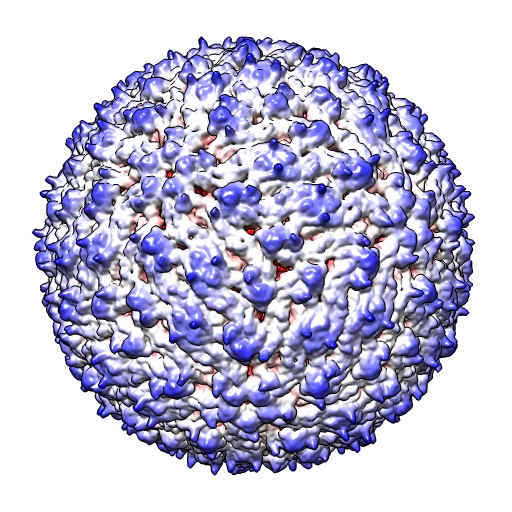
An approximation developed for video games called "screen space ambient occlusion" computes the darkening of recesses entirely on the GPU using the depth of each pixel. Here are some images using this technique from the PMV molecular viewer.
| 
| 
| 
|
| Normal lighting. Movie. | PMV ambient occlusion with default settings. Movie. | Black background. | PMV settings |
| 
| 
| 
|
| Scale 10, samples 50. Movie. | Blocky for most views | Ribbon | Surface |
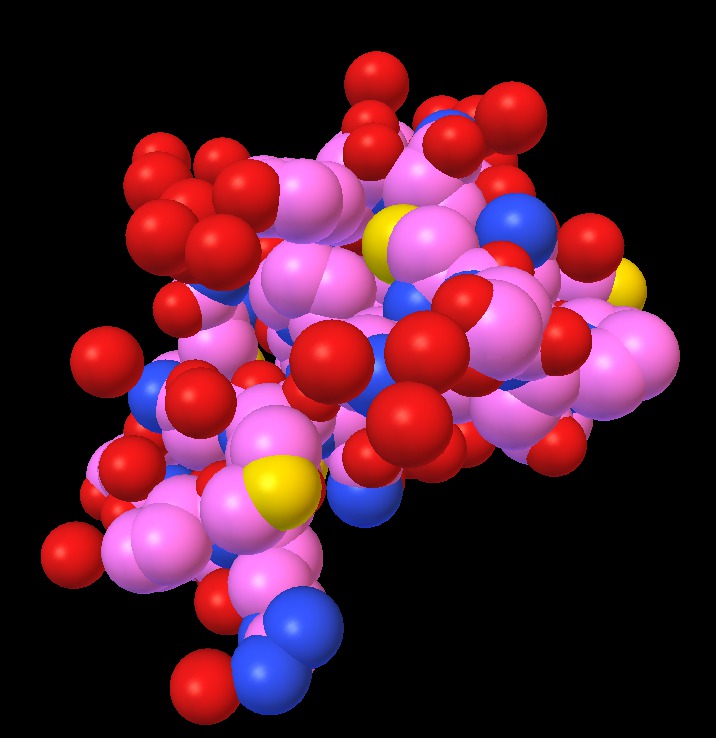
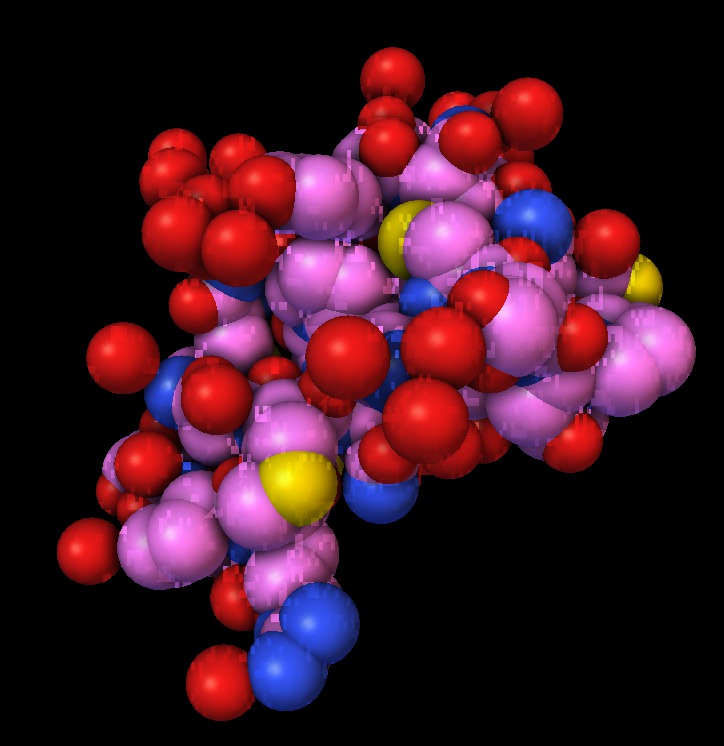
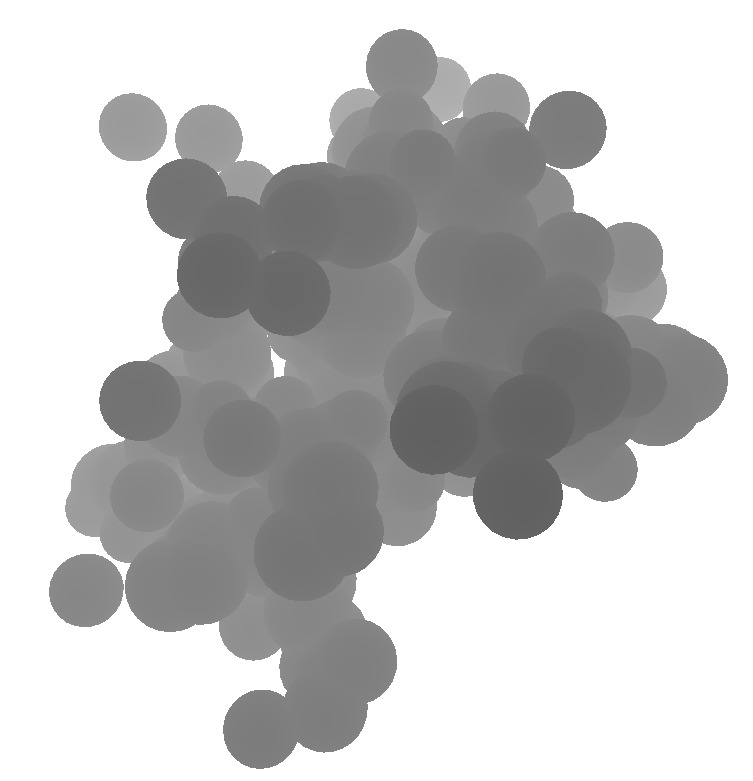
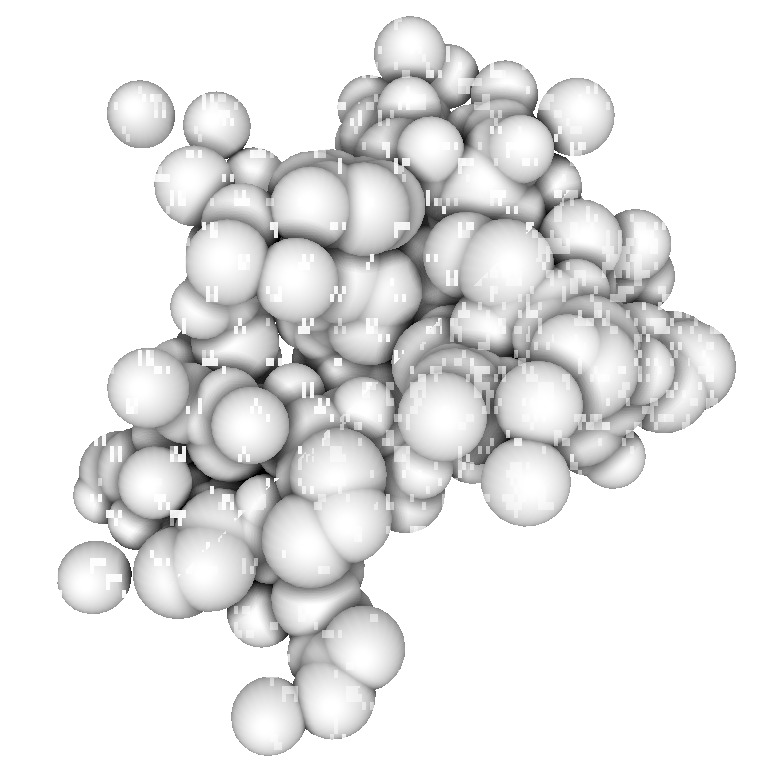
| 
| 
| 
| 
|
| 1a0m normal ligting. Movie. | 1a0m ambient occlusion. Blocky. Movie. | Close up | Depth map | SSAO modulation |
The blocky artifacts in PMV ambient occlusion prevent judging whether screen space ambient occlusion would have adequate quality for molecular visualization. The artifacts may be a graphics driver problem (tested on two different Mac Nvidia systems) or a PMV implementation bug. In addition to blockyness, the method produces undesirable halos around molecules on non-black backgrounds.
The PMV from MGLTools latest build (July 10, 2014) just made blocky dark areas. The current production release 1.5.6 (Feb 2012) gave better results shown in the images. Both require X windows on Mac. To enable SSAO I used the DejaVu dialog (shown by a toolbar button), Camera tab.
Frame rate for chaperonin 1aon 60,000 atoms sphere style was only 4.4 frames per second (60 in Chimera), apparently does not use instancing, and with ambient occlusion with default settings turned on the frame rate drops to 2.0 frames per second.
Rotating or zooming 1aon gave a constantly shifting multicolor blocky pattern with ambient occlusion enabled. May be a bug in PMV or a graphics driver issue (Mac 10.9.4, Nvidia GTX 680MX and Nvidia GT 650M). Same bad appearance on Mac laptop and desktop. Tried adjusting dozen parameters and switch options and none removed the blocky patterns. Blocky noise even appears scattered at random when no molecule is shown, indicating some sort of bug. Perhaps it is related to random sampling kernel orientation.
Screen space ambient occlusion is a very approximate method. Depths at each pixel are not sufficient information to judge occlusion in general. The ambient light reaching pockets does not depend on view angle, but the screen space method recalculates for each view and does depend on view angle. Sampling few depths near a pixel is sensitive exactly which points are sampled and blurring of the result is needed to reduce these artifacts. The blurring step causes the darkened regions to bleed for instance onto the background in PMV which is undesirable.
DisadvantagesMolecular graphics is mostly static scenes viewed from different angles and precomputing ambient occlusion might be simpler and better quality. The screen space method was developed for video games where dynamic scenes are much more common. The future of molecular graphics may use more dynamic scenes (morphs).
In order to assess the potential of the screen space method, it would be necessary to test a high quality implementation on example molecular systems. I don't know if a high quality implementation is available.
The density map methods for approximating ambient occlusion can be generalized to using a 3d texture in OpenGL which gives the ambient light reaching each point in space. The scene can then be rendered with brightness of objects modulated using this 3d texture. The above tests in Chimera and Hydra of the density map approach show that a very coarse grid (size 20 x 20 x 20) is adequate for a large molecule. Computing the ambient occlusion values to put in the 3d texture would not have to be density based. It could use a more accurate model that calculates where light can actually reach.
AdvantagesThe cases where ambient occlusion is useful, large space-filling views, are not the most common (atoms as spheres, molecular surfaces, EM map surfaces). Molecular ribbons with side chains are more often used. So handling a few special cases with a density map approach may get 80% of the benefit with simpler, more robust code, with more reliably high quality appearance.
| 
|
| QuteMol | Hydra |
QuteMol is a demonstration program showing ambient occlusion and edge highlighting on molecules developed in 2006. Development on it ended in 2007 (version 0.4.1). QuteMol shows high quality ambient occlusion and is interactive, at least 30 frames per second on 60,000 atom 1aon, taking about 3 seconds to load and display that molecule.
I tested the QuteMol 0.4.1 Mac Intel binary from 2007, and read the journal article to understand its methods.
It precomputes ambient occlusion for a molecule with atoms shown as spheres (or ball and stick) by calculating shadow maps in many (possibly hundreds) of directions on the GPU. It uses "imposters" for rendering spheres (and cylinders), meaning that a sphere is drawn as a square with the GPU discarding the points outside a circle, computing appropriate z values and normal vector in the fragment shader. Imposters are much faster than rendering triangulated spheres. In order to record the variation of the ambient occlusion lighting across each sphere they cover every sphere with small rectangular patches (4 to hundreds) and the rectangular patches are packed into large 2d textures. Mapping from the patch texture coordinates to and from the sphere points are used. There is a great deal of complexity handled in GPU programs for this texture mapping. The say that 15 shadow map directions per second could be handled with 60,000 atom 1aon on 2007 hardware. They did not say how many directions are needed for good quality. The qutemol.cfg file indicates 128 directions are used with relatively small 256x256 shadow map texture for each direction.
The Qutemol article also describes edge highlighting with variable thickness edges, thicker where the depth drop is greater. Also it describes halos that are transparent bright regions around each sphere. Also it describes capping clipped spheres on the GPU when using a near clip plane. All these techniques can be tested in their program.
Ambient Occlusion and Edge Cueing to Enhance Real Time Molecular Visualization Marco Tarini, Paolo Cignoni, and Claudio Montani IEEE TRANSACTIONS ON VISUALIZATION AND COMPUTER GRAPHICS, VOL. 12, NO. 5, SEPTEMBER/OCTOBER 2006
The basic QuteMol approach to computing ambient occlusion of saving the shading in textures and precomputing those textures is complex code. It does not handle molecular surfaces, density maps, ribbons, and doesn't generalize to arbitrary surfaces. In its favor it uses an essentially correct method for computing occlusion (sampling many light directions) as opposed to density approaches that give a length scale dependent result.
Could the basic idea of computing occlusion using a large number of light directions be used? This could be done on the CPU with Chimera mask command code for surfaces, and the same method could be adapted for spheres. It would me much faster on the GPU and could probably be done there, accumulating an occlusion value for each surface vertex or each sphere instance.
A basic difficulty of all ambient occlusion precomputation techniques is that they need to be recomputed whenever what is displayed changes. It will be necessary to detect any geometry changes. And the computation could be slow taking a second whenever geometry changes, making it undesirable to use by default.
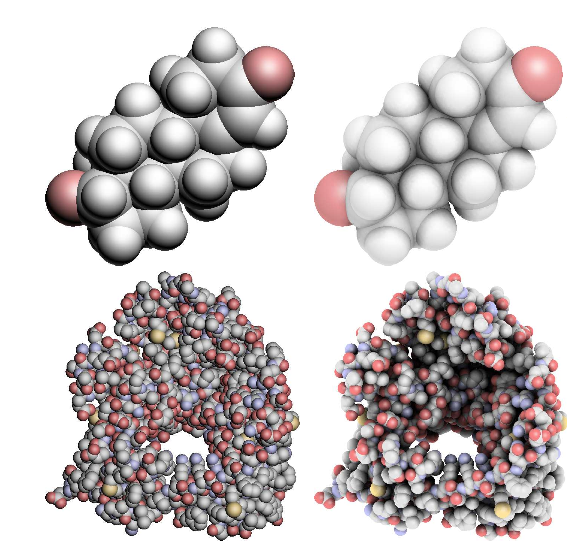
| ||
| QuteMol article fig. 8. Direct illumination (left) and ambient occlusion (right) for small (above) and medium (below) molecules. Above: testosterone (49 atoms) and, below, porin (2219 atoms). | ||
|
While ambient occlusion lighting looks great on big molecules (100,000 atoms) shown with atom spheres, its depth effect is much less impressive on smoother objects, like small molecules, or low resolution EM maps. This is clear in QuteMol article figure 8, where the 49 atom molecule in the top row has good depth without ambient occlusion (left), while on the second row with 2219 atoms, the depth is poor without ambient occlusion.
What stands out is that direct lighting (ambient, diffuse, specular as done by Chimera and other programs) looks horrible on pebbly surfaces such as thousands of atom spheres. It gives no sense of depth. Ambient occlusion depth looks fine on such pebbly surfaces.
If conventional direct lighting worked as well on pebbly surfaces as it does on smooth surfaces then there would be only a small benefit of using the difficult to compute ambient occlusion. Can direct lighting be fixed to work on pebbly surfaces as well as it works on smooth surfaces?
An extreme example suggests that a pebbly surface is going to look not much different in its shading from looking at a flat surface face on. Viewing a sawtooth surface with the view direction parallel one tooth edge, then the observed surface normal vectors are all in the same direction as if this were a flat plane parallel the other tooth edge.
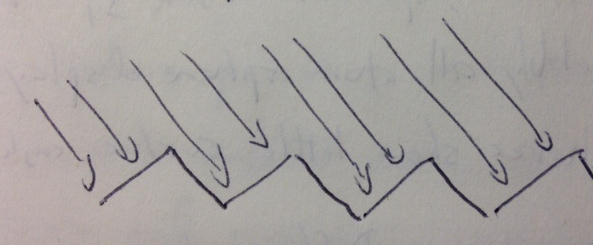
| 
|
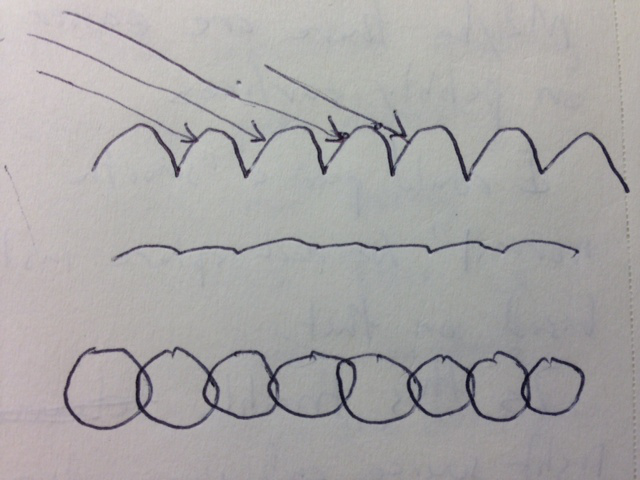
Looking at a flat plane of packed spheres also suggests that the normal ambient, diffuse, and specular lighting won't create much variation of brightness bassed on the angle of the plane to the viewing or light directions. Tests in Chimera show that diffuse lighting without ambient or specular shows little angle dependent brightness variation on spheres (1aon), nor does specular without diffuse show much brightness variation on the spheres, and of course ambient depends on neither view direction or light directions so it shows no variation in brightness.
So it does not appear that lighting based on surface normal, view direction and light direction can give brightness variation on pebbly surfaces to indicate the orientation of the pebbly plane relative to the view direction. But it would be simple to enhance the lighting if a smoother surface normal was provided to the graphics shader. This normal could be obtained as a gradient of a density map of the atoms computed at atom centers to give one smooth normal per sphere, or at molecular surface vertices. The smooth normal would contribute another diffuse lighting term. The actual normal would still be used in addition, otherwise the pebblyness of the surface would be invisible.
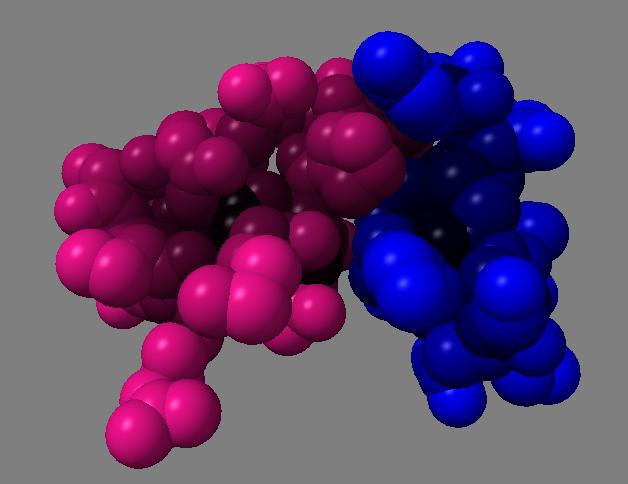
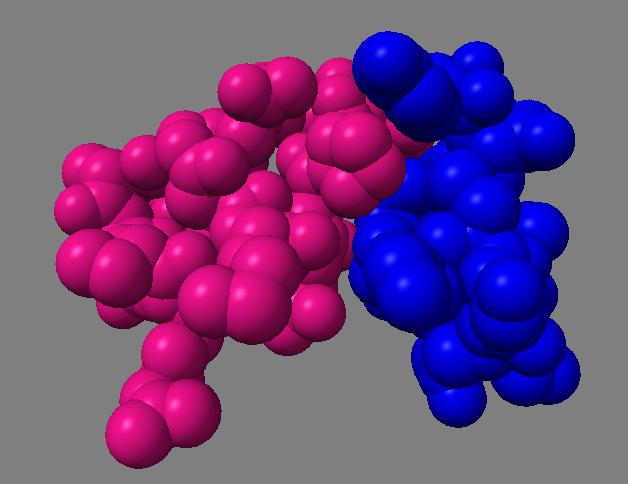
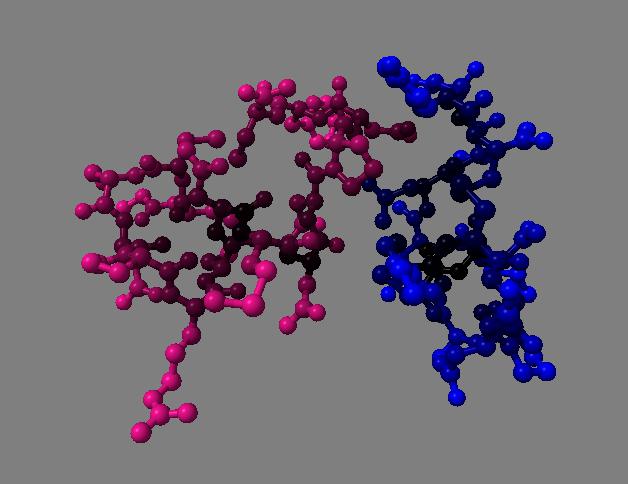
For 250 atoms (pdb 1a0m) single color spheres looks worse than spheres that have gradations of color. Ball and stick ambient occlusion doesn't help depth perception the darkened crease between each ball and stick.
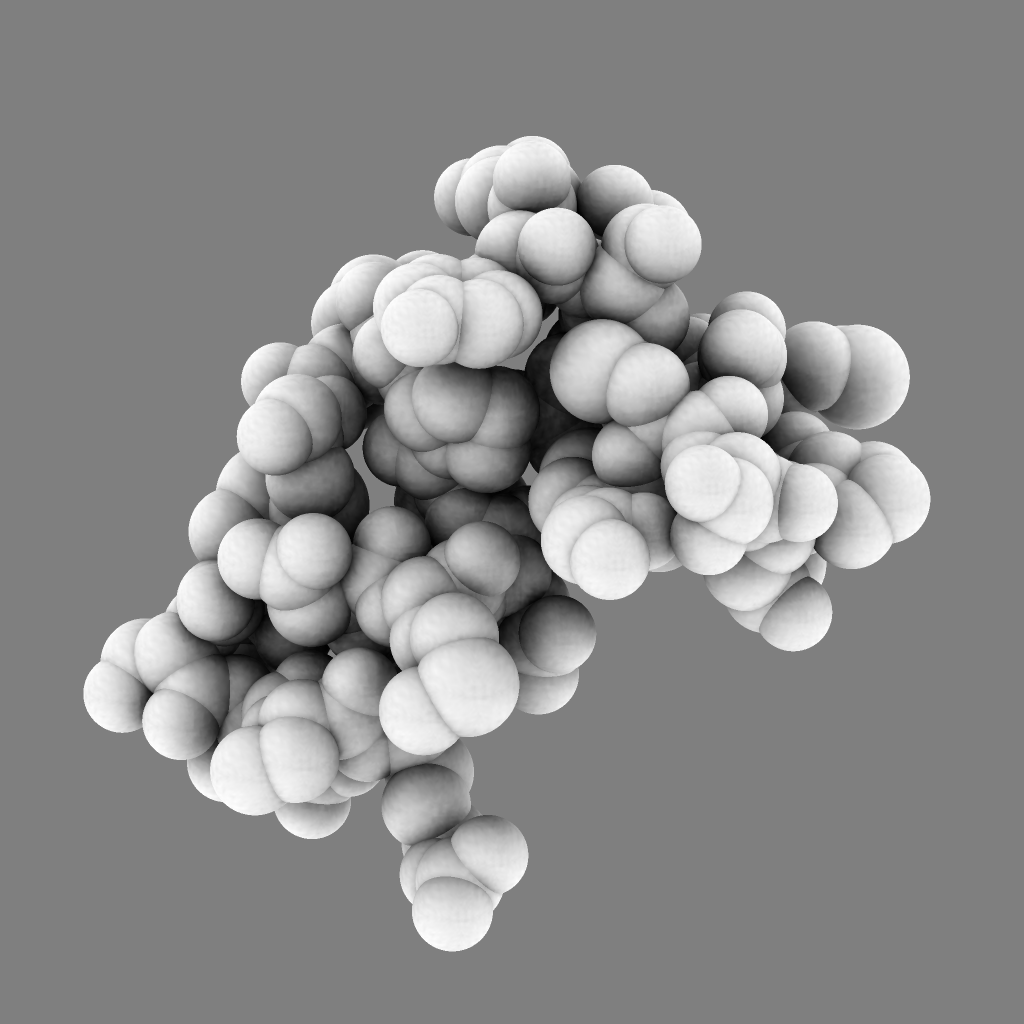
| 
| 
| 
| 
|
| QuteMol | Hydra density map method | QuteMol direct lighting | QuteMol ambient occlusion | Hydra ambient occlusion |
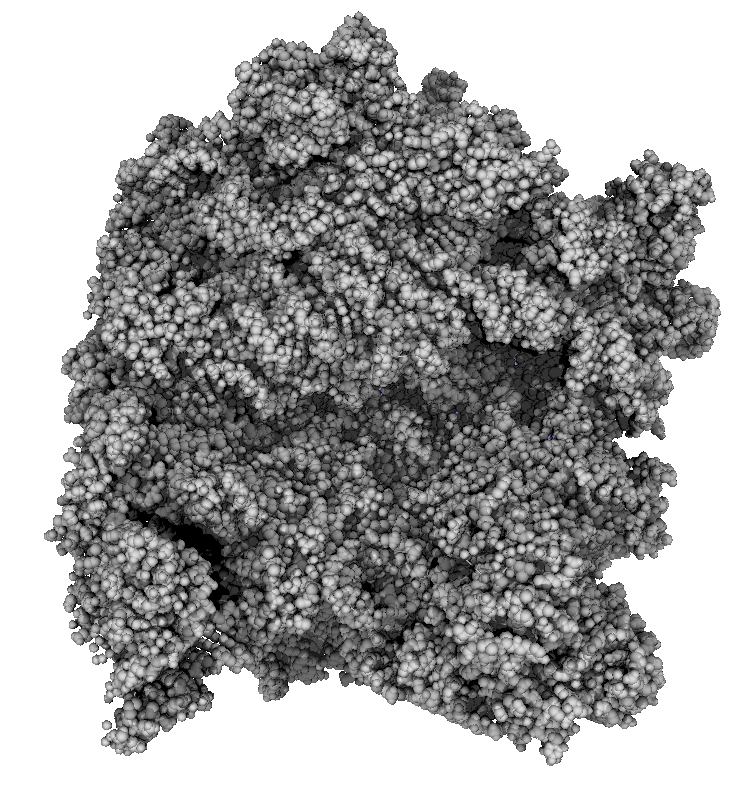
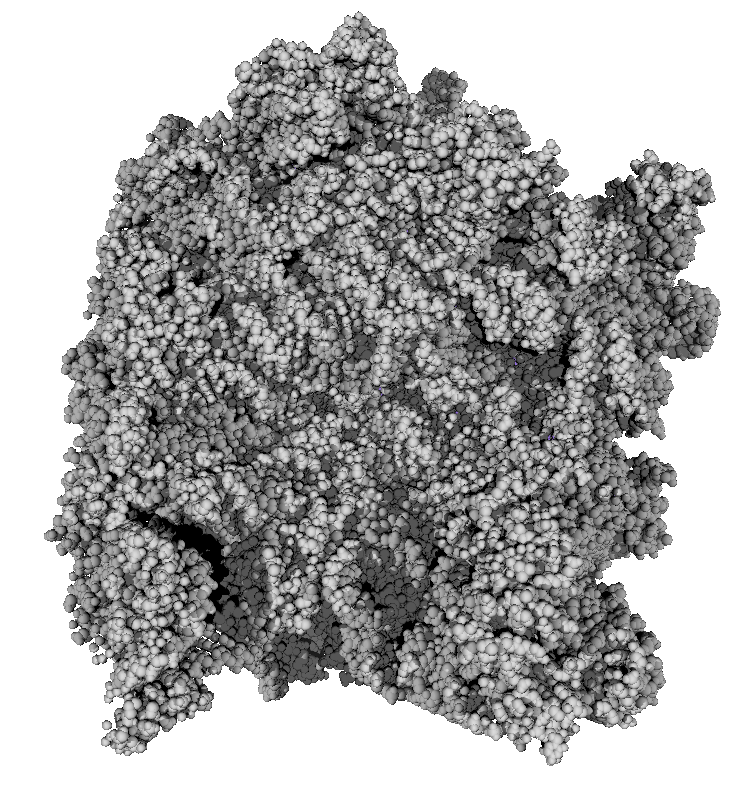
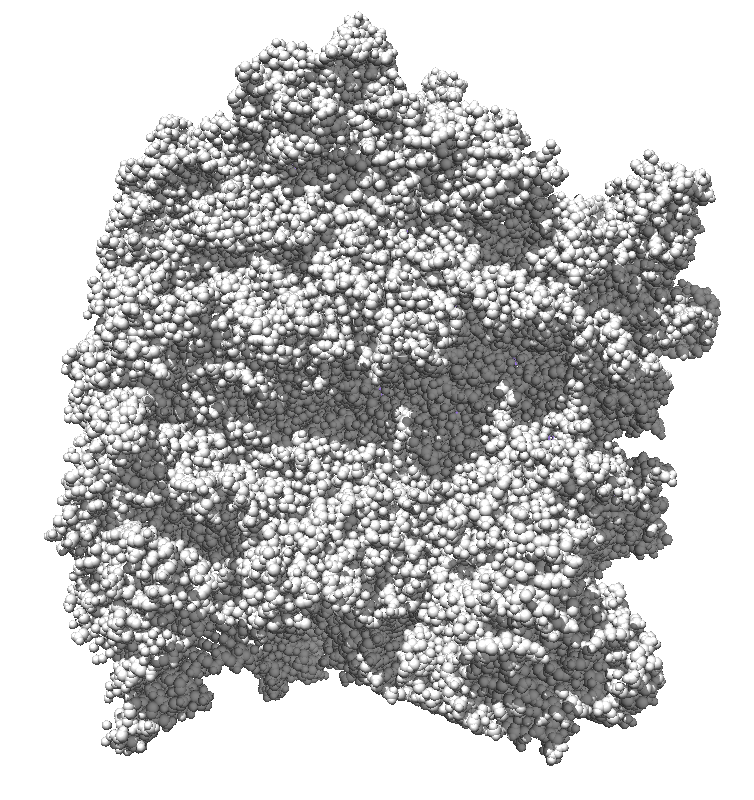
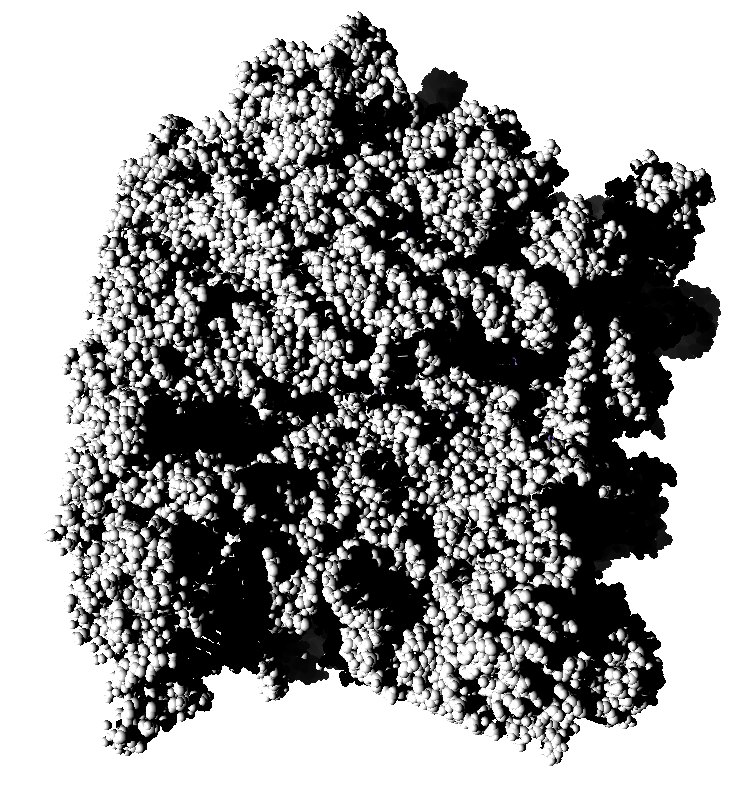
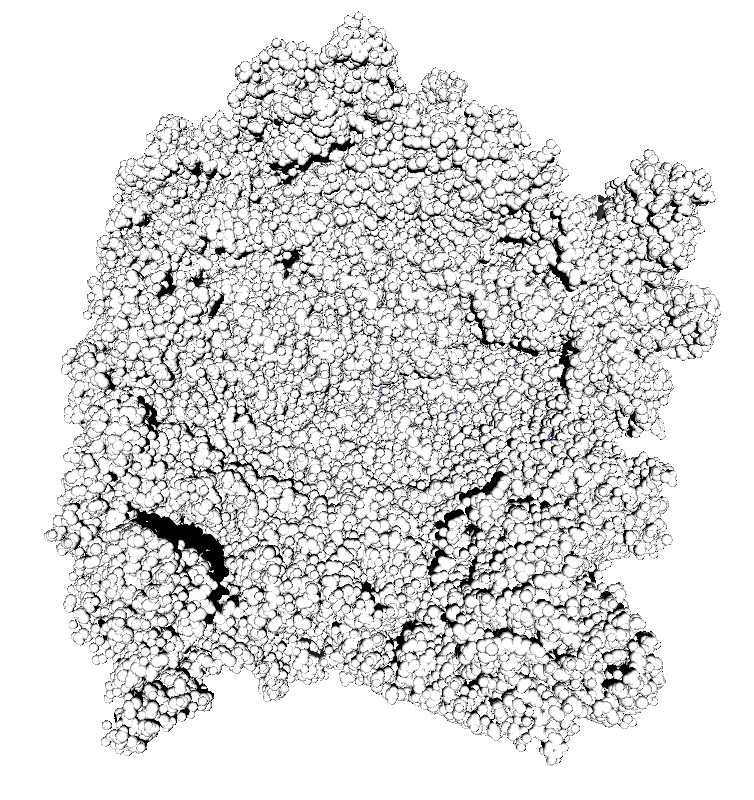
How well do 2, 3, 4, 6 or more directional lights casting shadows convey depth compared to ambient occlusion? Could test this using Chimera which can only produce shadows for one light by saving multiple images of the same view with different light directions and averaging them together. (ImageMagick can do the average "convert -average *.png output.png"). If lights from fixed to the camera are used then the shadows will move around in possibly a distracting way. Instead could use shadow lights that rotate with the model so that no motion of the shadows would be seen.
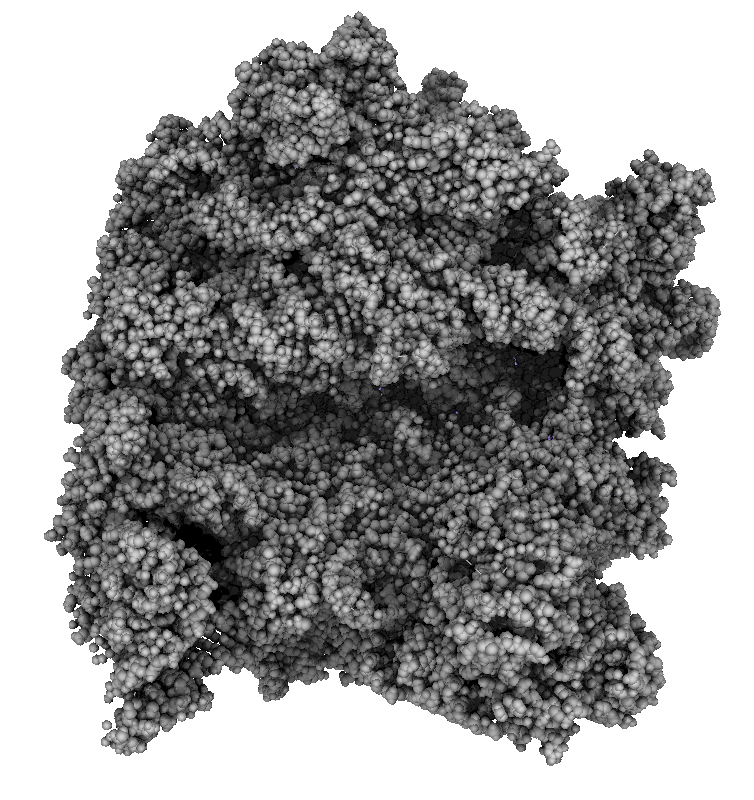
| 
| 
| 
|
| 9 shadows blended | 5 shadows | 4 shadows | 4 shadows diagonal |
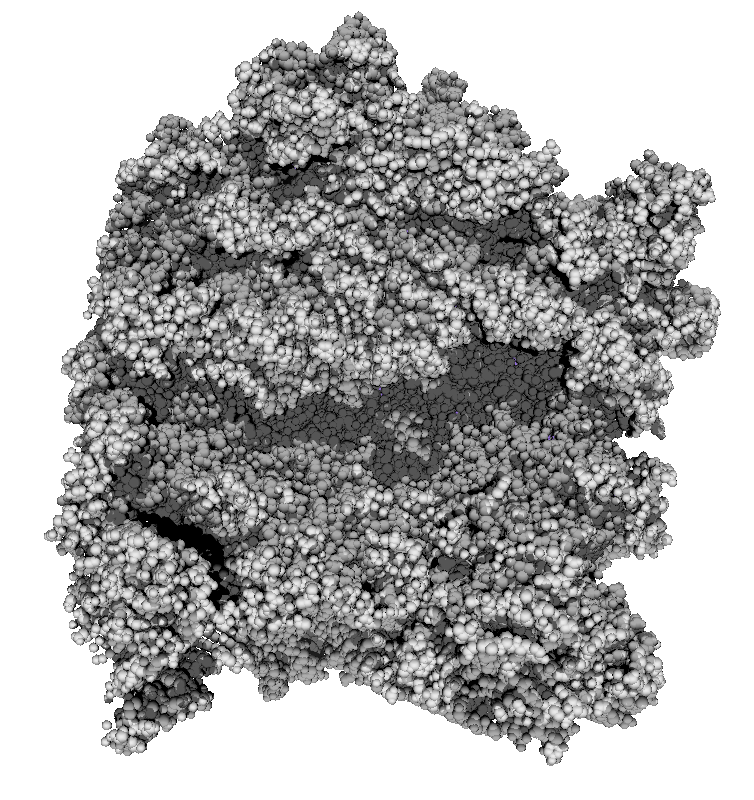
| 
| 
| 
|
| 3 shadows vertical | 3 shadows horizontal | Standard Chimera lighting | Standard 1 shadow. |
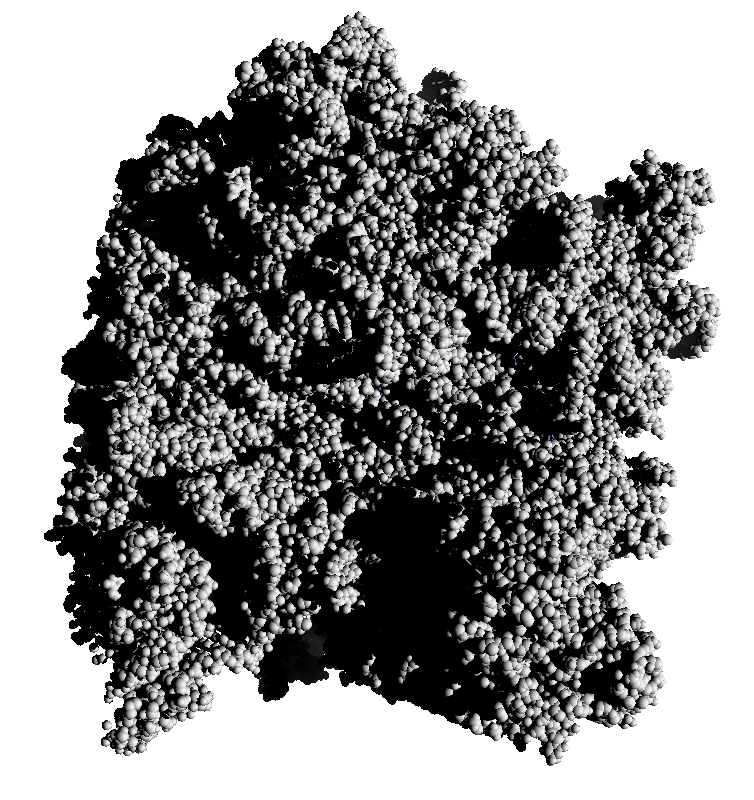
Subsets of the 9 images below were averaged to produce the multishadow images above. Light directions were vx = -1/0/1, vy = -1/0/1, vz = 1.

| 
| 
|
| 
| 
|
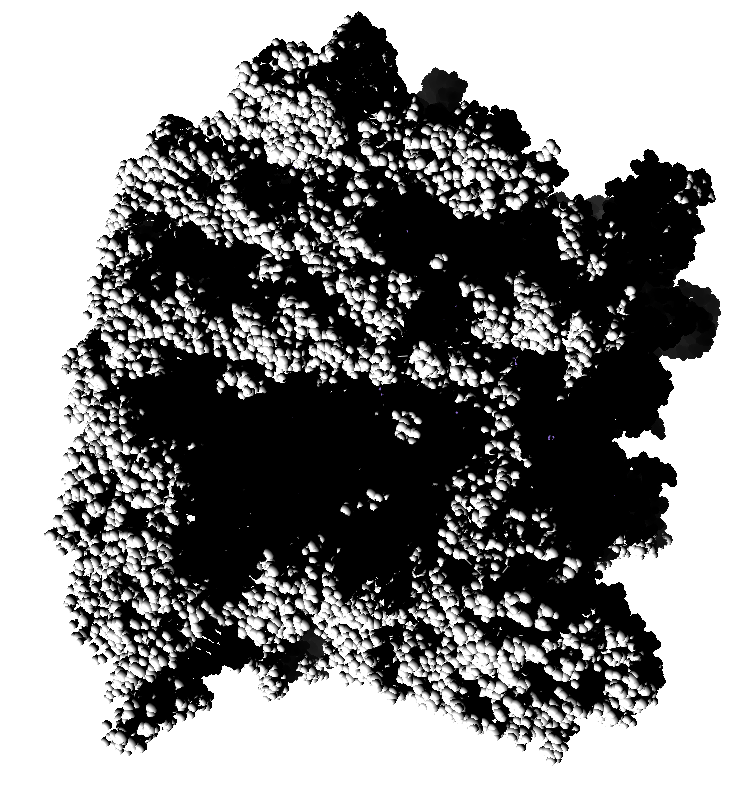
| 
| 
|