
 |
|
Custom Colors in MidasPlus
 JPEG
version (73KB), TIFF version
(387KB)
JPEG
version (73KB), TIFF version
(387KB)
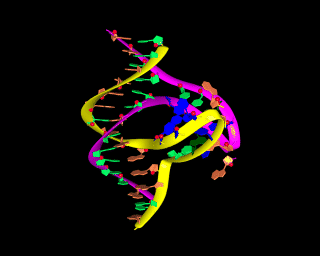
This is an image of a model of Slip-Loop DNA (see below for details on the structure of this model). The thing to note in this image are the colors. These colors were custom made by the user using the "colordef" command. Especially note the pastel green and orange colors, which do not belong to the predetermined set of 65 MidasPlus colors.
In MidasPlus, any combination of red, green, and blue can be mixed to make your own colors.
command: colordef light_green .3 .9 .3To use this color,
command: color light_green :45-55 (This will color residues 45 through 55 light green, the custom color)To make a color lighter, add equal parts of the remaining colors. For example. To make pink, add equal parts of green and blue to a full strength red:
pink 1.0 .4 .4To make a color darker, do not use the full strength of that color.
dark_red .7 0.0 0.0The only real way to learn how to get colors you like is to experiment a little. All numbers need to be between 0 and 1. Other notable colors, as guidelines, are:
yellow 1.0 1.0 0.0 orange 1.0 .5 0.0 magenta 1.0 0.0 1.0 cyan 0.0 1.0 1.0Also, on Silicon Graphics workstations, /usr/sbin/cedit can be used to interactively experiment with color combinations. Cedit reports color components in the range 0-255, so it is necessary to divide cedit's values by 255 for use with the colordef command.
This is a novel hypothetical type of folding for nucleic acids. It may form for DNA sequences with short direct repeats (about 5-6 base pairs long), when one strand of DNA is shifted relative to another. Two loops, formed as a result of such shift, have a potential to form tertiary base pairs. Such a structure may be stabilized in circular DNA by a superhelical stress. Also, SLS may be formed for RNA with a special sequence.
Calculations were performed with the DNAminiCarlo program (N.B. Ulyanov, A.A. Gorin V.B. Zhurkin) which is currently under continuing development in Tom James' lab. ©2004 The Regents, University of California; all rights reserved.