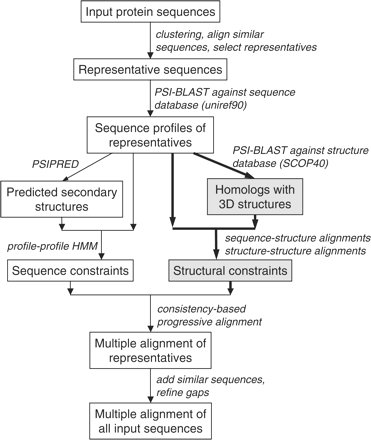
|
SABmark-twi (209/10667) |
SABmark-sup (425/19092) |
|
(# mult alignments/# pairwise ref alignments) |
| Method |
Q-score
(max ≈ 0.71) |
GDT-TS |
Q-score
(max ≈ 0.87) |
GDT-TS |
|
| PROMALS3D (D + S) |
0.602 |
0.264 |
0.805 |
0.417 |
| PROMALS3D (F + S) |
0.555 |
0.220 |
0.779 |
0.390 |
| PROMALS3D (T + S) |
0.540 |
0.249 |
0.766 |
0.412 |
| PROMALS3D (D + F + S) |
0.611 |
0.256 |
0.812 |
0.414 |
| PROMALS3D (D + T + S) |
0.603 |
0.264 |
0.805 |
0.421 |
| PROMALS3D (F + T + S) |
0.595 |
0.251 |
0.800 |
0.413 |
| PROMALS3D (D + F + T + S) |
0.616 |
0.260 |
0.812 |
0.420 |
| 3DCoffee (D + S) |
0.574 |
0.252 |
0.802 |
0.421 |
| 3DCoffee (SAP + S) |
0.553 |
0.222 |
0.786 |
0.390 |
| Expresso webserver |
0.508 |
0.206 |
– |
– |
| PROMALS3D (D/2 + S) |
0.475 |
0.198 |
0.716 |
0.364 |
| 3DCoffee (D/2 + S) |
0.261 |
0.100 |
0.573 |
0.294 |
| 3DCoffee (D/2 + SAP) |
0.255 |
0.095 |
0.572 |
0.289 |
| --------- sequence-only methods --------- |
| PROMALS |
0.393 |
0.154 |
0.665 |
0.336 |
| SPEM |
0.326 |
0.124 |
0.628 |
0.318 |
| MUMMALS |
0.196 |
0.081 |
0.522 |
0.278 |
| ProbCons |
0.166 |
0.058 |
0.485 |
0.246 |
| MAFFT-linsi |
0.184 |
0.070 |
0.510 |
0.264 |
| MUSCLE |
0.136 |
0.056 |
0.433 |
0.233 |
| T-Coffee |
0.134 |
0.048 |
0.429 |
0.223 |
| ClustalW |
0.127 |
0.057 |
0.390 |
0.221 |
| --------- structure-only methods --------- |
| MUSTANG |
0.550 |
0.230 |
0.779 |
0.404 |
| PROMALS3D (D) |
0.594 |
0.252 |
0.802 |
0.415 |
|