|
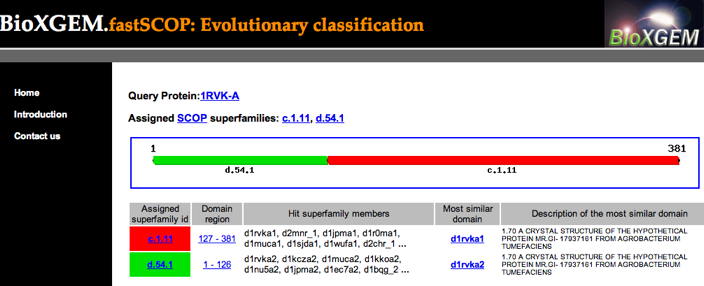
I entered 1rvk chain A, then waited about 10 seconds...
|
|
|
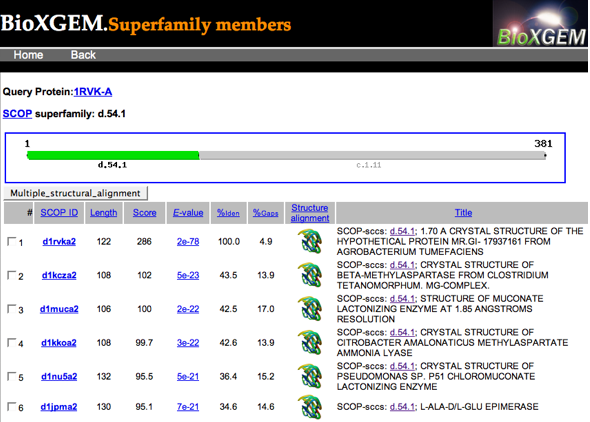
Then I clicked on the link for the N-terminal domain region, which
gives a list of hits in the assigned superfamily.
There must be some cutoff for what gets listed, because when I
searched with two different enolase superfamily members, I got different
numbers of hits listed
for the SCOP "Enolase N-terminal domain-like" superfamily.
|
|
|
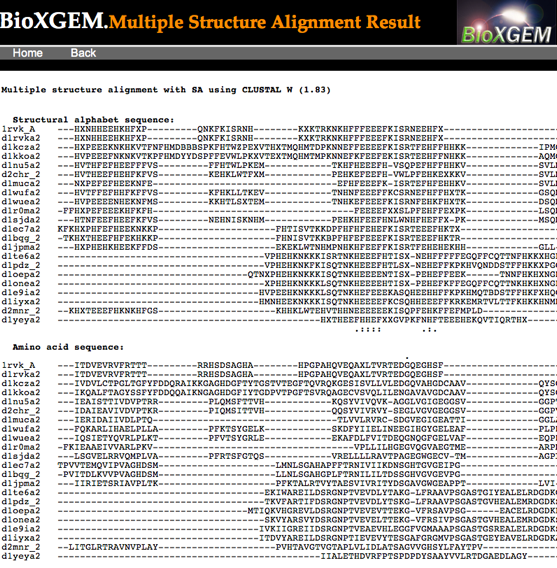
Clicking a structure icon gives an image and Chime-plugin view of a
pairwise superposition and an option to download them in PDB format.
Instead, I checked all the boxes (there was an option to do that at the
bottom) and clicked the "Multiple_structural_alignment" button...
|
|
|
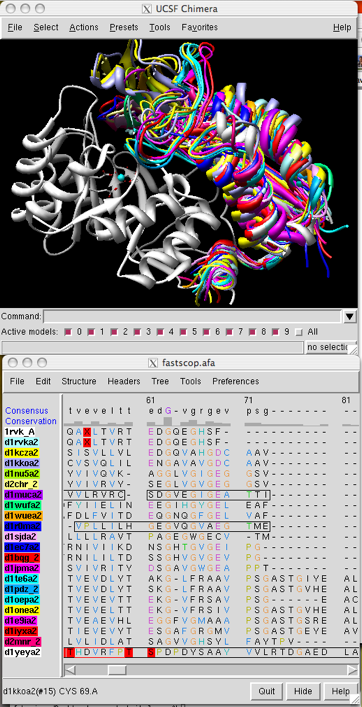
I cut and pasted the amino acid sequence alignment into a text file
and edited it into an aligned
fasta file. I opened the fasta alignment in Chimera,
auto-loaded all the structures (since they were named by
PDB and SCOP domain IDs), and matched them using the alignment.
|
|





