
seen by electron microscopy.

| 
|
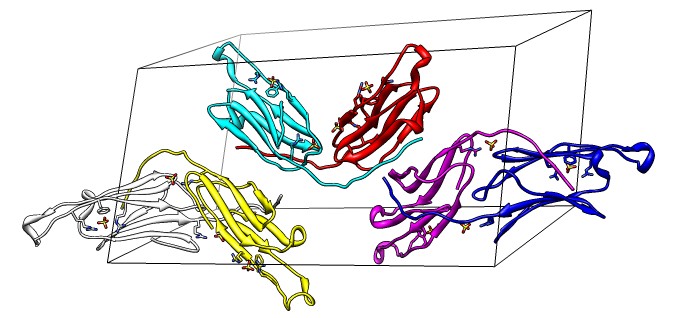
| 24 copies of alpha crystallin for oligomer seen by electron microscopy. | Human alpha crystallin is 2 beta sheets. |
| 
| 
|
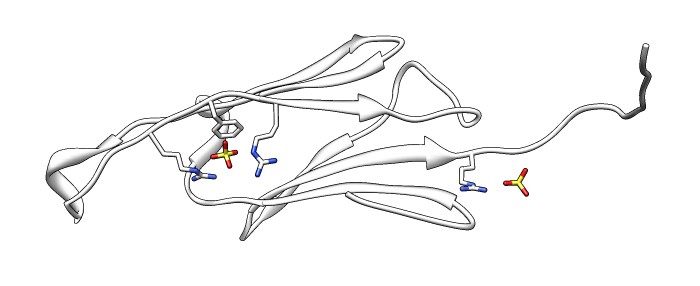
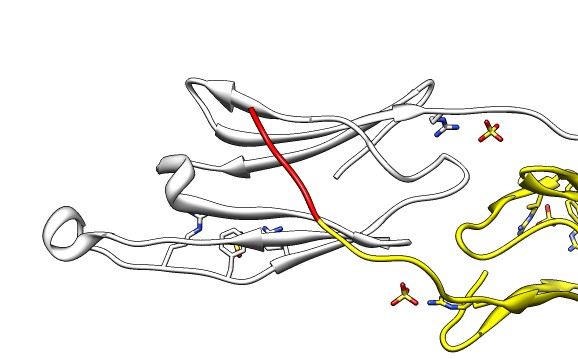
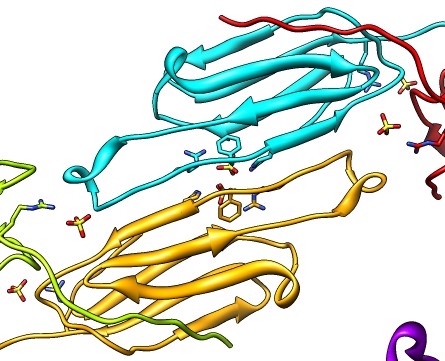
| Various crystal structures show several biological dimerization modes. | C-terminal tail binds to edge of beta-sheet. Binds in 180 degree flipped orientation too. Binding sequence is palindrome. | Side-by-side beta sheet dimer. |
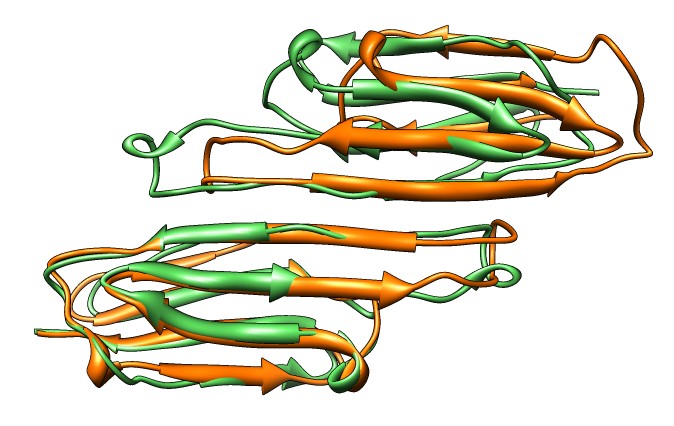
| 
| |
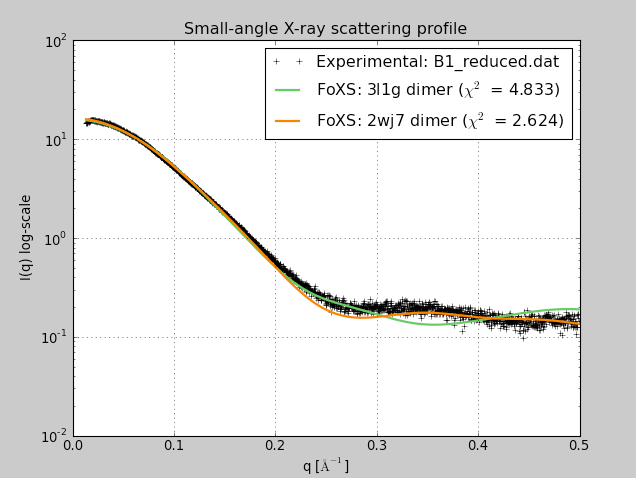
| Beta-sheet dimer observed with 3 different registrations in crystals and NMR. Some literature suggest all are present allowing different oligomers to form. | Small-angle x-ray scattering in solution shows only one registration is present at physiological conditions (NMR registration). |
|
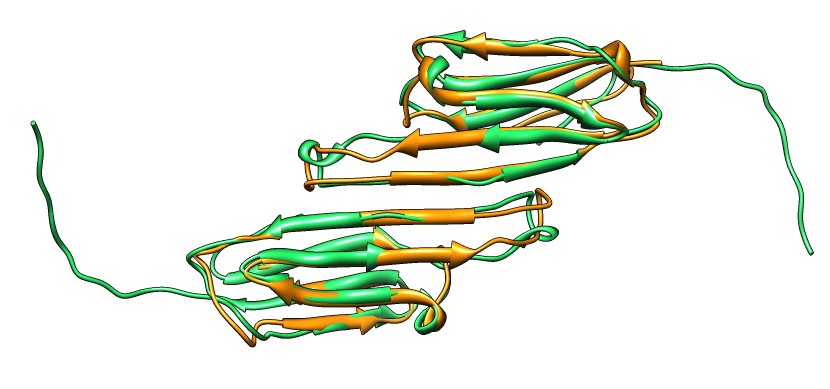
| Shifted human dimer with C-terminal tails for building 24-mer. |
Fit dimer to map. Determine map symmetry and replicate fit dimer. Adjust tails by matching to crystal tail dimer and using energy minimization. Show hydrophobicity SES surfaces. Hide C-terminal tails to show very hydrophobic groove -- oligmer believed to disassemble to bind unfolded proteins -- exposes these grooves. Show M jannashii sHSP octahedral symmetry, 1shs. Morph between 1shs and the 3l1g fit created in the demo. Mention that higher resolution 9A EM map and fit is about to be published -- will be interesting to see how far off our quick model is.