Tom Goddard
November 19, 2007
This year's progress and coming projects.
Try to give some depth on our new EM tomography efforts, and breadth of our undertakings with many current users.
Chimera density map capabilities are used primarily by researchers using single particle EM reconstruction to study molecular assemblies.
Map display and analysis tools developed over past 6 years focused on single particle EM with primary collaborator Wah Chiu and his NCMI resource center.
Survey of capabilities in Guide to Volume Data Display. Display, fitting, carving, and measuring maps and handling large multimeric atomic models (viruses, ribosomes, filaments, ...).
Estimate 500-1000 EM Chimera users based on email support requests from ~100 distinct users per year.
Chimera has been used for exploring EM tomograms (calculated from tilt series of single specimens) although we did not develop tools specifically for this use until a year ago.
Why add EM tomography capabilities to Chimera?
Strong encouragement from collaborators Manfred Auer (LBNL, sterociliar bundles), Wah Chiu (National Center for Macromolecular Imaging), Wolfgang Baumeister (MPI, cellular organization), Yifan Cheng (UCSF, clathrin vesicles), John Sedat (UCSF, chromosomes), Gary Ren (UCSF, low density lipoprotein cholesterol packaging).
Growing literature of tomography applications to molecule-scale architecture, where Chimera atomic model capabilities can be used. Here are a few examples that used Chimera for EM tomography (all completed before we added specialized tomography support).
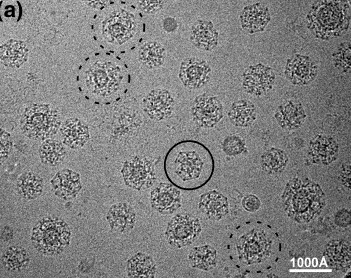
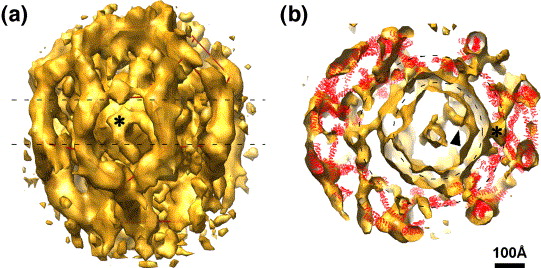
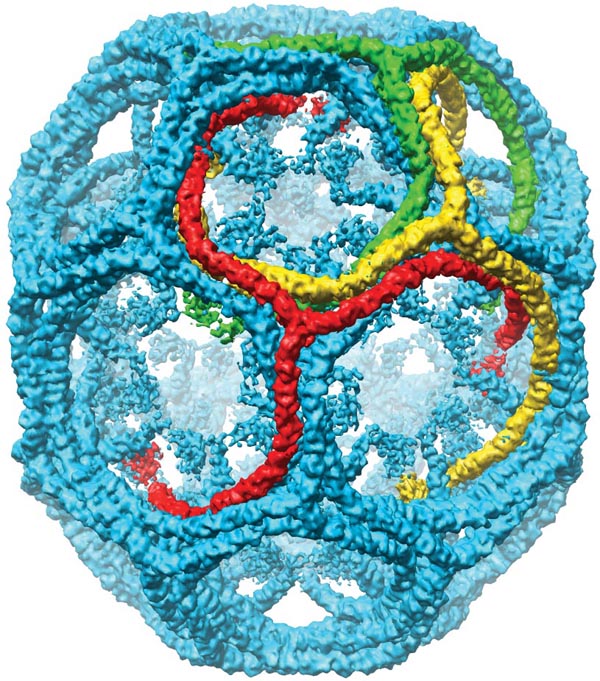
Clathrin vesicles, Yifan Cheng.
Cheng Y, Boll W, Kirchhausen T, Harrison SC, Walz T.
| 
| 
|
| In vivo clathrin cages have many topologies. | Modeling a cage from tomogram. | In vitro prepared clathrin allows averaging. |
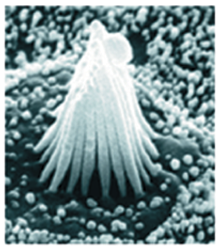
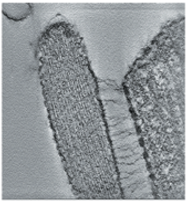
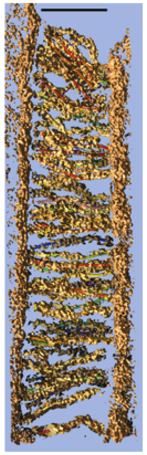
Stereocilia molecular linkers (unpublished), Manfred Auer lab.
| 
| 
| 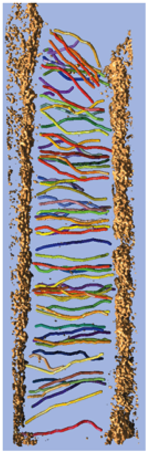
| Traced links composed of a few molecules. Scale bar 100 nm. |
| SEM of hair bundle | Tomogram section of tip links. |
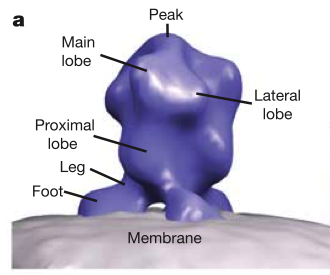
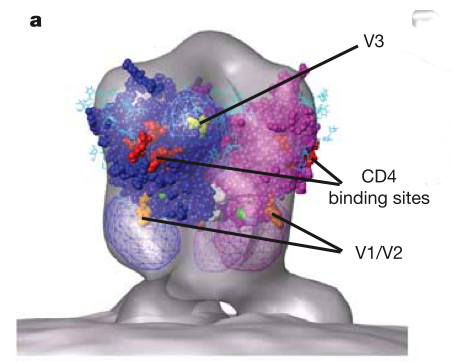
HIV glycoprotein spikes, Roux lab.
Zhu P, Liu J, Bess J Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH.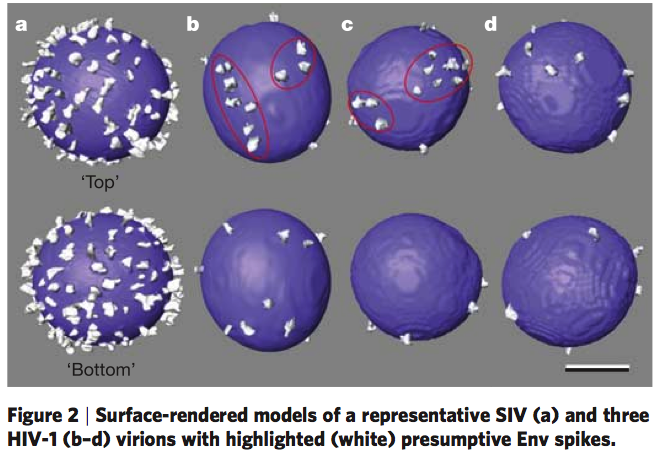
| 
| 
|
Carboxysomes, Michael Schmid (co-director of NCMI).
Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM.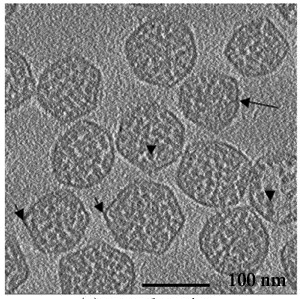
| 
|
| Carboxysomes encapsulate RiBisCO in cyanobacteria. | Averaging 21 tomogram particles with diameter ~100 nm suggests arrangement of capsid molecules. |
Herpes virus portal, Wah Chiu lab.
Chang JT, Schmid MF, Rixon FJ, Chiu W.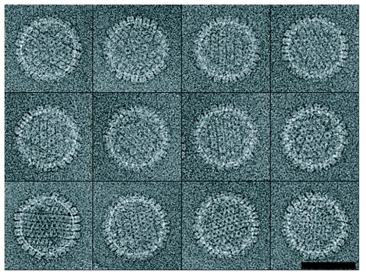 Herpes simplex virus. Tomographic sections of several particles. | 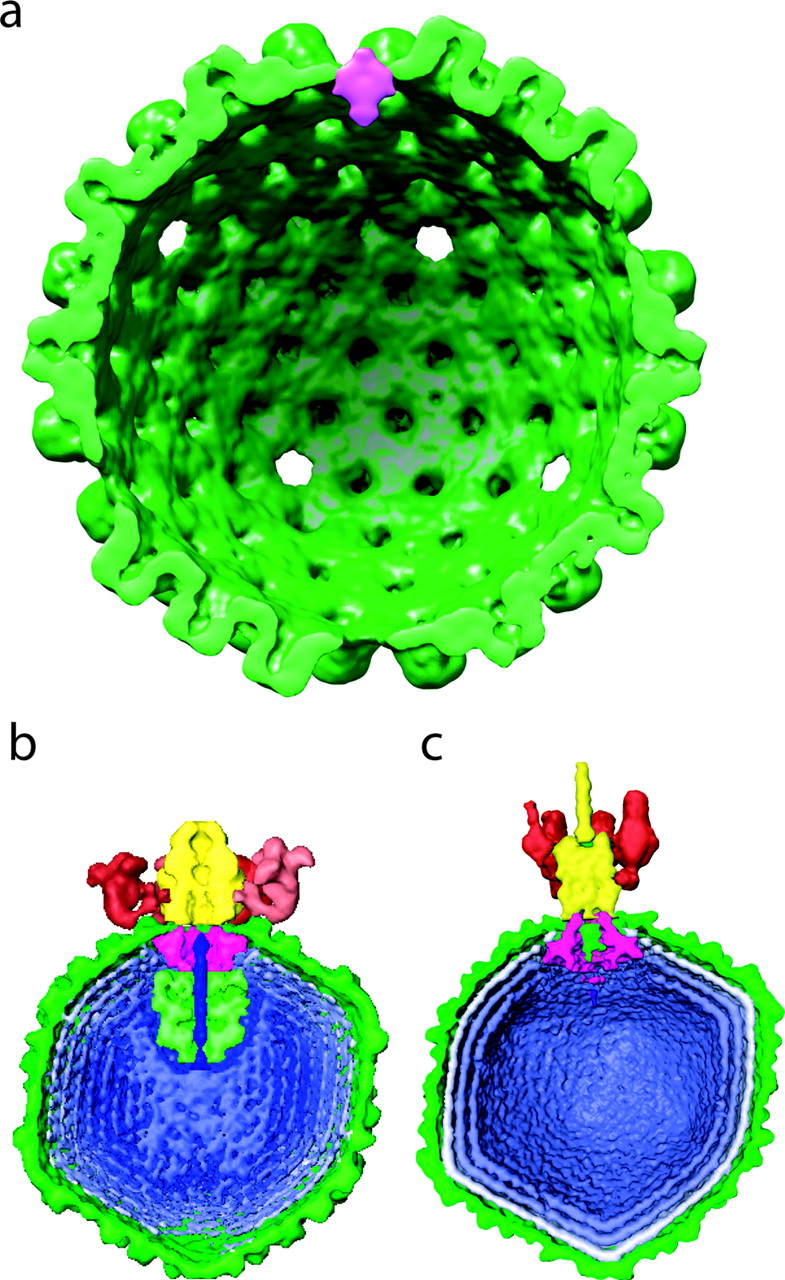
| One of 12 vertices occupied by a portal of nearly equal mass to 11 pentons. Tomographic reconstruction used to identify portal vertex and 13 particles averaged. Large Epsilon 15 and P22 virus portals shown for comparison below. |
Viewing arbitrary cross-sections by interpolating data on a plane.
|
| 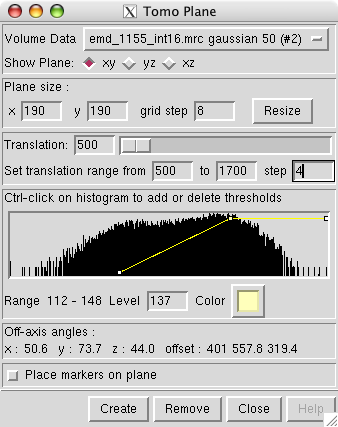
Single HIV particle, Gaussian smoothed to 5 nm to improve visiblity of
core. Conincal core oriented at 45 degrees in xy plane, and axis is
tilted out of xy plane.
|
Tomo Plane Chimera extension developed by Karin Gross and Christoph Best
in the Baumeister lab at Max Planck Institute for Biochemistry.
| |
|
Another way to looking at arbitrary cross-sections is to resample the
data on a rotated grid oriented and sized with the mouse. The rotated submap
can be saved for use in other software that supports plane by plane
segmentation of only xy planes (e.g. IMOD or Amira).
| |
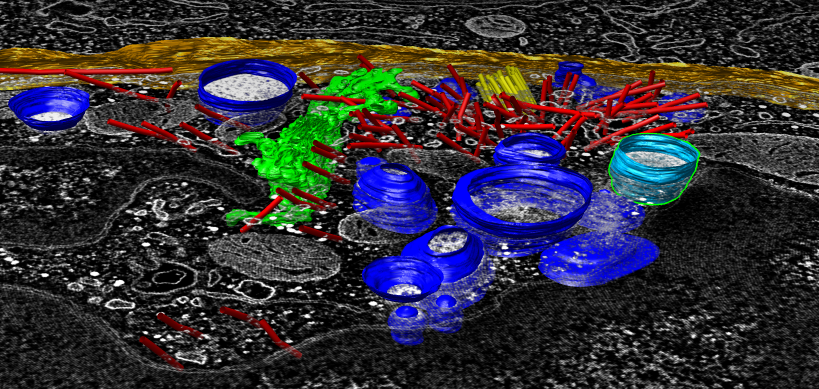

|
Tomogram of human cytotoxic T-cell killing neighbor cell (beyond orange cell membrane). Segmented lysosomes (blue), microtubles (red), centriole (yellow), golgi (green), cell boundary (orange) done with separate IMOD software. Chimera capabilities
|
Tomograms are large files. They require efficient handling for interactive visualization. Typical dimensions 2048 by 2048 by 100 (400 Mbytes). The latest detectors produce 4096 by 4096 images and gigabyte size tomograms.

|
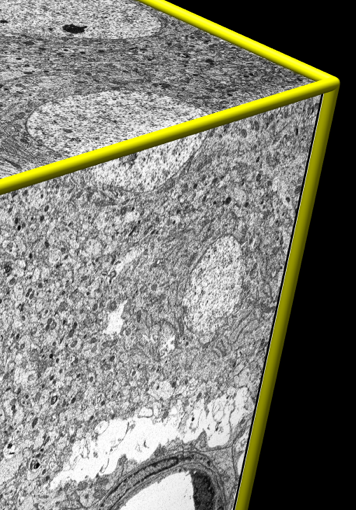
|
Rat cortex. 50 x 50 x 100 um. 8 Gbytes, 2048³ volume. Voxel 25 x 25 x 50 nm. One voxel fits ~2 ribosomes. |
Problems
Tricks for large map files.
Tried all these using HDF file format. This is part of a collaboration with the National Center for Macromolecular Imaging (Wah Chiu), the EM Data Bank (EBI, Kim Henrick), and the PDB (Helen Berman, John Westbrook), to design a new high performance EM map format. Other capabilities we have implemented:
Example speedups:
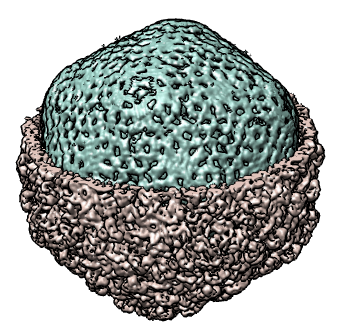
Show masking out a shell of a virus using gaussian filtered surface. Phage K1E (EMD 1334). 50Å Gaussian smoothing used to define two surfaces for separating protein capsid from packed genome.

| 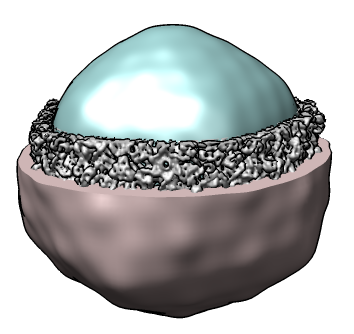
| 
|
Show saving map symmetries in hdf file, sym command contact option, and detect clashes.
|
Improved fitting with symmetric molecule copies.
|
|
Would like to fit in a brief showing of face capping. Quality-of-implementation improvement.
Made filament building script for Ben Keshet, amyloid Advised Alexander Solodukhin on fitting 3 domains in kinase map Helped Angel Paredes with map display graphics driver problems Debugged ccp4 map origin problem, n*start heuristic, Walter Novak Helping Dhiraj with selecting atoms near portal of channel map Discussing ellipsoid molecule models with Bernard Heymann (Bsoft) Advised Gabe Landers on helixhunter vrml output Checked quadro 900 xgl freezes reported by Hernando Sosa Helped Terry Lang with volume interpolation Added graphics benchmark scores from Dave Konerding, Damir Perisa, Gabor Papai... to web page Wrote script to build path tracer helix for Evan Sadler Looked at writing imagic-5 volume file reader, Michael Schatz Wrote simple python script to make map images, Eduardo Sanz Garcia Wrote python script to interpolate map at atom values, Terry Lang Made fit map in map no gui python script for Gabor Papai Advised Hirofumi Suzuki of chimera features for em navigator web site Made imod file reader handle contours and point sets, David Morgan Helped Gabor Papai with morphing between differently normalized maps Advised Andrea Rossi on chimera crash with firegl graphics, bug 3797 Helped Rich Kaufman, save pdb, ribbon shown, save displayed atoms only Installed / tested eman chimera extension pdb to mrc, Thomas Marlovits Helped Janet Iwasa with vrml output of dotted pseudo-bonds Helped Michel Dedeo with model motions for animation Wrote mx keyboard shortcut to move markers to map maxima, Navnit Mishra Advised on figure scale and mrc/brix/o alignment questions, C Akey Advised Mazdak Radjainia about fitting and path measuring Advised Mazdak Radjainia on merging maps Helped Christopher Akey with chimera crashes, large ribosome sessions Helped Ettore Panizon with myosin image for controluce web site Added fit map axis point output for finding sym axes, Albion Baucom Providing fix for saving pdb relative to map, Bernard Heymann Answering chimera quest.: Pu Qian, Chad Smith, Jeff Triffo, Yifan Cheng Fixed tom_em volume data session restore bug, Stephan Nickell Made script to save pdb coords for block of unit cells, Brady Bernard Helped Randy Heiland with chimera problem using numpy and numeric Advised on multiple volume dialogs and save relative, Batsal Devkota Packaged fit map in map for Gabor Papai Added coloring of surfaces by norm of volume gradient, David Morgan Tried virus electrostatic coloring for 1ny7 for Xiaobin Fan
Asked Steve Ludtke, Pawel Penzcek, Matt Dougherty about hdf map format compatibility between eman and chimera Discussed wii remote inteface to chimera with Adam Marko Discussed segmentation of light microscopy with Sebastian Haase Helped Jeff Triffo with bad opengl fog / volume rendering behavior Helped Ed Egelman with unexpected blue ribbon coloring with pov-ray Advised Ed Egelman on crashes with large pdb models and maps Phone discussion with Jeff Triffo, vtk for surface mean curvature Showed path tracer and other chimera tools to Yifan Cheng Advised Yao Cong on lapack chimera issue and 2-d contours (Chiu group) Richard Newman visited for day, met with me and Tom Ferrin Trained Richard on various map presentation image tricks Arranging visit to nat. center for x-ray tomography, Carolyn Larabell Answered volume menu, model closed programming questions, Karin Gross Sent Kim Henrick suggestions for EMDB voxel size fix and image gallery Advised Richard Newman on handling incorrect ccp4 header nsymbt value Answered email request M Lombardia, C Lawson, P Natarajan Explained counting atoms outside density contour, Gary Ren Created example motion monitoring code for Karin Gross Added dpi setting to viper multiscale image script, Padma Natarajan Added image dpi to viper em script, Padma Natarajan Wrote accelerator to print radial average density for virus maps Investigated Matt Shawkey, Hildur P. bird feather em and advised Helped Jeff Triffo with copying volume settings and tracing surfaces Met Kim Henrick, Helen Berman at eman workshop to discuss em masks Advised Ed Egelman on surface capping for showing filament lumen Made euler angle marker set rotation script, Jeff Triffo Tested Wei Zhang's volume interpolater and made it a separate extension Created code to send map data to chimera, Stephan Nickell Helped Matt Shawkey with volume fourier interpolation script Created radial coloring nogui script for Gabe Lander Phone meeting on new em file format, Matt Dougherty + 7 others (dec) Discussed fitting multiple helix pieces to chromosomes, Heather McCune Showed Gary Ren chimera map features for 2 hours Created mesh to molecule command for Gabe Lander Helped Mike Marsh with stereo focal plane setting Met Chris Best (Max Planck Institute) and Mike Hohn (LBL) and discussed Chimera for em tomography Helped Matt Dougherty with chimera stereo questions and 64-bit linux Read ear hair bundle linkers manuscript from Manfred Auer Helped Steve Ludtke with isosurface color bug, likely graphics driver
made sym command use volume symmetries and have range cutoff added chimera hdf: subsamples, x-planes, symmetries, rotations, compression, and multiple maps (masks) per file separated ccp4 and mrc file readers, better origin handling resample data and save rotated orientation solid rendering of skewed data added volume command cleaned up python volume api hdf eman file reader improved initial transfer function improved volumetric opacity model z-plane volume interface many improvements to volume user interface (step adjust, hide/show, multiple initial volume colors...) tiff image stack reader cancel volume read, progress reports read only volume subregion into memory multiple histograms in volume dialog thick silhouette edges imod contours using path tracer markers mac crash reporting fetch by id fo-fc EDS xray maps fetch by id pqs c++ headers in distributions movie roundtrip option movie raytracing movie supersampling faster movie capture using ppm format volume/area auto-update electrostatics volume opens surface color Studying serial block face sem rat cortex data Wii remote experiments Requested space group in ccp4 eds maps Chimera release party (june) Added 2007 chimera bug reports to graphics driver bugs web page contour face capping request from Lars Liljas for T4 baseplate images for textbook Made web page of chimera plugins written by others Review vnc use for chimera collaboration (group meeting) invert map, unsigned 8-bit mrc map accelerators Made contouring work with larger volumes Updated numeric to numpy Put cellular complexity, site visit, and bioimage talks on web Added netcdf support for precomputes subsamples in same file as data Made fit model/map use only the displayed subregion of the base map Made routine and shortcut to double map along z axis for microtubule Added windowsize command to chimera distribution for Elaine Meng Created chimera change log and feature request web pages
We promote Chimera use by the EM community by giving demonstrations and hands-on tutorials at meetings:
and through publications.
Possible things to show:
Major theme is new capabilities for EM tomography.
Also many enhancements in functionality and usability for higher resolution
maps and models of assemblies.
Show HIV spike tomography averaging and Yifan's clathrin vesicles to
connect tomography and single particle EM.
Immune cell tomography demo
IMOD mesh reader
TomoPlane extension
Plane display
Extract subregions not aligned with data axes
HDF volume file capabilities
symmetry matrices for microtubule
save rotated orientation
sub-samples for large data sets
x-slices for large data sets
compression of masks, CTL masks, centriole
multimap saving for masks
unsigned 8-bit data values
Symmetric copies command with range cutoff
Detect clashes -- use with microtubule neighbor monomers
Can I work rat cortex data into demonstration? Will be pressed for time.
Can I show Gaussian filter? Showed at last adcom with HIV particles.
If I can split IMOD connected pieces objects and measure volume
that would be good to show. Could mention dendrite spines case.
Fetch EDS maps and PQS biounits interesting but don't fit into demo.
Don't plan on showing Wii.
Should I mention or list many quality of implementation improvements?
Should list them and probably not talk about any one. So they know where
our time is going.
Should I list help I provided for user requests? Good to list and quantify
a bit but not discuss any in detail.
Should say something about collaborations. Mention morph map from Wei Zhang.
Were viper em map radial color images in past year? Yes, showed to adcom.
List meetings with Chimera EM/assemblies presentations (5 since last adcom).
Coming year focus areas: 1) integrating multiscale seamlessly into Chimera
to make it visible and simpler to attract more users. 2) Using, creating
volume masks. List examples (mmcif, low-res surfaces for single molecule
use, masks for virus layers).
Other new stuff (taken from progress web page, March - Sep 2007):
made sym command use volume symmetries and have range cutoff
added chimera hdf: subsamples, x-planes, symmetries, rotations, compression,
and multiple maps (masks) per file
separated ccp4 and mrc file readers, better origin handling
resample data and save rotated orientation
solid rendering of skewed data
added volume command
cleaned up python volume api
hdf eman file reader
improved initial transfer function
improved volumetric opacity model
z-plane volume interface
many improvements to volume user interface (step adjust, hide/show,
multiple initial volume colors...)
tiff image stack reader
cancel volume read, progress reports
read only volume subregion into memory
multiple histograms in volume dialog
thick silhouette edges
imod contours using path tracer markers
mac crash reporting
fetch by id fo-fc EDS xray maps
fetch by id pqs
c++ headers in distributions
movie roundtrip option
movie raytracing
movie supersampling
faster movie capture using ppm format
volume/area auto-update
electrostatics volume opens surface color
More stuff (from 2007+2006 log files):
Curr opin review vis software for assemblies.
vis density maps paper came out jan 1, 2007, submitted july 2006
Studying serial block face sem rat cortex data
Wii remote experiments
Requested space group in ccp4 eds maps
Chimera release party (june)
Added 2007 chimera bug reports to graphics driver bugs web page
contour face capping
request from Lars Liljas for T4 baseplate images for textbook
Made web page of chimera plugins written by others
Review vnc use for chimera collaboration (group meeting)
invert map, unsigned 8-bit mrc map accelerators
Made contouring work with larger volumes
Updated numeric to numpy
Put cellular complexity, site visit, and bioimage talks on web
Added netcdf support for precomputes subsamples in same file as data
Made fit model/map use only the displayed subregion of the base map
Made routine and shortcut to double map along z axis for microtubule
Added windowsize command to chimera distribution for Elaine Meng
Created chimera change log and feature request web pages
Several user help calls per week (Might list representative dozen, person/task)
Made filament building script for Ben Keshet, amyloid
Advised Alexander Solodukhin on fitting 3 domains in kinase map
Helped Angel Paredes with map display graphics driver problems
Debugged ccp4 map origin problem, n*start heuristic, Walter Novak
Helping Dhiraj with selecting atoms near portal of channel map
Discussing ellipsoid molecule models with Bernard Heymann (Bsoft)
Advised Gabe Landers on helixhunter vrml output
Checked quadro 900 xgl freezes reported by Hernando Sosa
Helped Terry Lang with volume interpolation
Added graphics benchmark scores from Dave Konerding, Damir Perisa, Gabor Papai... to web page
Wrote script to build path tracer helix for Evan Sadler
Looked at writing imagic-5 volume file reader, Michael Schatz
Wrote simple python script to make map images, Eduardo Sanz Garcia
Wrote python script to interpolate map at atom values, Terry Lang
Made fit map in map no gui python script for Gabor Papai
Advised Hirofumi Suzuki of chimera features for em navigator web site
Made imod file reader handle contours and point sets, David Morgan
Helped Gabor Papai with morphing between differently normalized maps
Advised Andrea Rossi on chimera crash with firegl graphics, bug 3797
Helped Rich Kaufman, save pdb, ribbon shown, save displayed atoms only
Installed / tested eman chimera extension pdb to mrc, Thomas Marlovits
Helped Janet Iwasa with vrml output of dotted pseudo-bonds
Helped Michel Dedeo with model motions for animation
Wrote mx keyboard shortcut to move markers to map maxima, Navnit Mishra
Advised on figure scale and mrc/brix/o alignment questions, C Akey
Advised Mazdak Radjainia about fitting and path measuring
Advised Mazdak Radjainia on merging maps
Helped Christopher Akey with chimera crashes, large ribosome sessions
Helped Ettore Panizon with myosin image for controluce web site
Added fit map axis point output for finding sym axes, Albion Baucom
Providing fix for saving pdb relative to map, Bernard Heymann
Answering chimera quest.: Pu Qian, Chad Smith, Jeff Triffo, Yifan Cheng
Fixed tom_em volume data session restore bug, Stephan Nickell
Made script to save pdb coords for block of unit cells, Brady Bernard
Helped Randy Heiland with chimera problem using numpy and numeric
Advised on multiple volume dialogs and save relative, Batsal Devkota
Packaged fit map in map for Gabor Papai
Added coloring of surfaces by norm of volume gradient, David Morgan
Tried virus electrostatic coloring for 1ny7 for Xiaobin Fan
Asked Steve Ludtke, Pawel Penzcek, Matt Dougherty about hdf map
format compatibility between eman and chimera
Discussed wii remote inteface to chimera with Adam Marko
Discussed segmentation of light microscopy with Sebastian Haase
Helped Jeff Triffo with bad opengl fog / volume rendering behavior
Helped Ed Egelman with unexpected blue ribbon coloring with pov-ray
Advised Ed Egelman on crashes with large pdb models and maps
Phone discussion with Jeff Triffo, vtk for surface mean curvature
Showed path tracer and other chimera tools to Yifan Cheng
Advised Yao Cong on lapack chimera issue and 2-d contours (Chiu group)
Richard Newman visited for day, met with me and Tom Ferrin
Trained Richard on various map presentation image tricks
Arranging visit to nat. center for x-ray tomography, Carolyn Larabell
Answered volume menu, model closed programming questions, Karin Gross
Sent Kim Henrick suggestions for EMDB voxel size fix and image gallery
Advised Richard Newman on handling incorrect ccp4 header nsymbt value
Answered email request M Lombardia, C Lawson, P Natarajan
Explained counting atoms outside density contour, Gary Ren
Created example motion monitoring code for Karin Gross
Added dpi setting to viper multiscale image script, Padma Natarajan
Added image dpi to viper em script, Padma Natarajan
Wrote accelerator to print radial average density for virus maps
Investigated Matt Shawkey, Hildur P. bird feather em and advised
Helped Jeff Triffo with copying volume settings and tracing surfaces
Met Kim Henrick, Helen Berman at eman workshop to discuss em masks
Advised Ed Egelman on surface capping for showing filament lumen
Made euler angle marker set rotation script, Jeff Triffo
Tested Wei Zhang's volume interpolater and made it a separate extension
Created code to send map data to chimera, Stephan Nickell
Helped Matt Shawkey with volume fourier interpolation script
Created radial coloring nogui script for Gabe Lander
Phone meeting on new em file format, Matt Dougherty + 7 others (dec)
Discussed fitting multiple helix pieces to chromosomes, Heather McCune
Showed Gary Ren chimera map features for 2 hours
Created mesh to molecule command for Gabe Lander
Helped Mike Marsh with stereo focal plane setting
Met Chris Best (Max Planck Institute) and Mike Hohn (LBL) and
discussed Chimera for em tomography
Helped Matt Dougherty with chimera stereo questions and 64-bit linux
Read ear hair bundle linkers manuscript from Manfred Auer
Helped Steve Ludtke with isosurface color bug, likely graphics driver
10/27/2006 Advisory committee meeting
11/15/2006 ncrr renewal site visit
12/13-15/2006 Attended workshop on visualizing cellular complexity in Munich
biophysical society meeting, baltimore march 5-6, booth presentation
eman workshop chimera tutorial (march 14-17)
chimera demo and tutorial at uc davis cryo-em meeting (May 4), Holland Cheng
nramm workshop nov 14, 2007
Debugged sparky 3114 on vista
Fixed sparky to eliminate gcc 4.0 compiler warnings, Jack Howarth
Advised on sparky fedora core 4 startup problem, Mark Kelly
Look at experimental features page for new stuff.
Look at EM Technology remove group meeting slides for possible reuse.
Should I make a pitch for cellular and tissue level tomography that
is not at molecular scale? It is exciting but a stretch. Basic idea
would be that we have built up enough EM vis tools that they have value
on there own without molecular models for exploratory use in cell/tissue
tomography. Tell them this is a possibly crazy idea -- I have not decided
if it is crazy or not. Current developments are motivated my molecular
scale tomography, but an exciting frontier may be this EM resolution
cell and tissue visualization. More study of existing software is needed
to see if Chimera is well positioned to fill this role. Don't make the
pitch -- but do show CTL and rat cortex data. Discuss possibility of
non-moleucule scale applications only if committee raises it.
Illustrate large map capabilities. Would be nice to show rat cortex.
Chunking helps accessing pieces for masking small regions.
Plane by plane on 4K by 4K tomogram (16 - 64 Mbytes per plane) is
slow (plane per second) without subsample map.