Keren Lasker
Tom Goddard
February 23, 2009
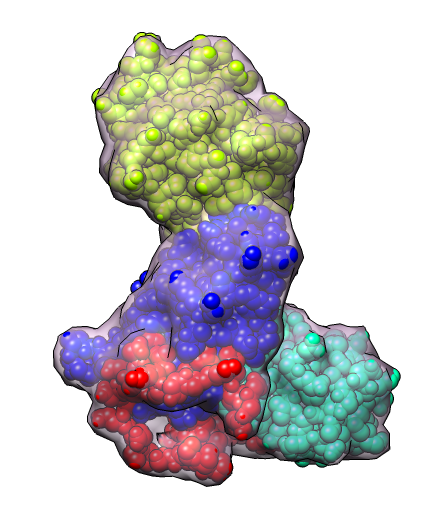
Illustration of fitting atomic models into EM map using MultiFit from Sali lab in combination with Chimera.
Other cases allow specifying restraints for which proteins contact which others and for allowing MultiFit to determine best protein locations on graph.

| 
| 
|
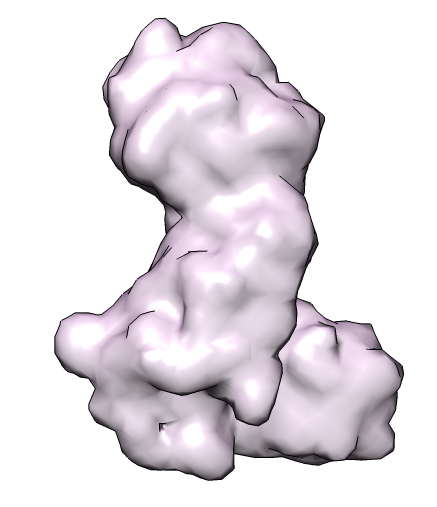
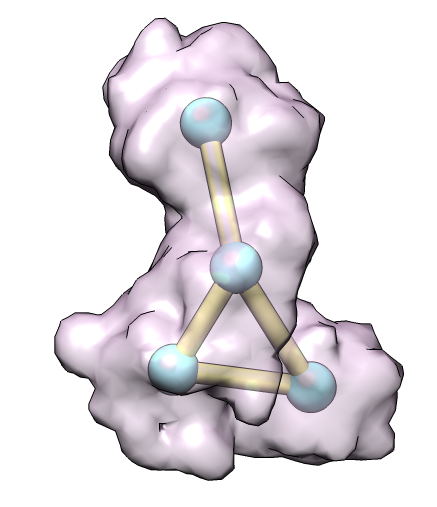
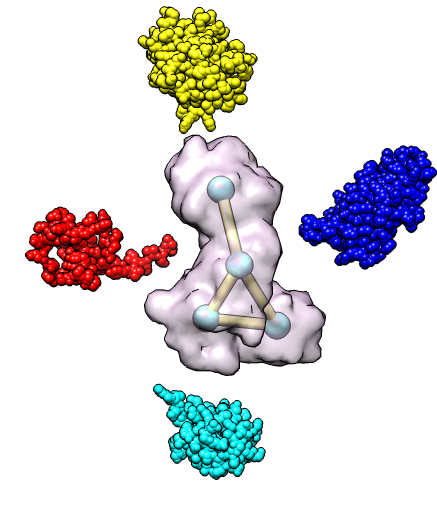
| 4 protein nuclear pore targeting complex (simulated map 9 A from 1z5s). | Graph partition of map using k-means algorithm. | Crystal structures for fitting into map. |

| 
| |
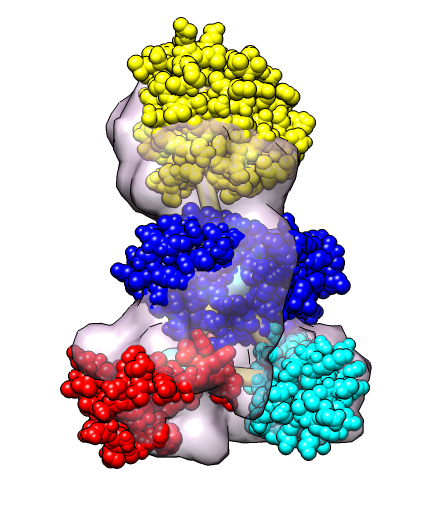
| Hand-placed proteins are associated with closest graph nodes. | Computational optimization by MultiFit program. |