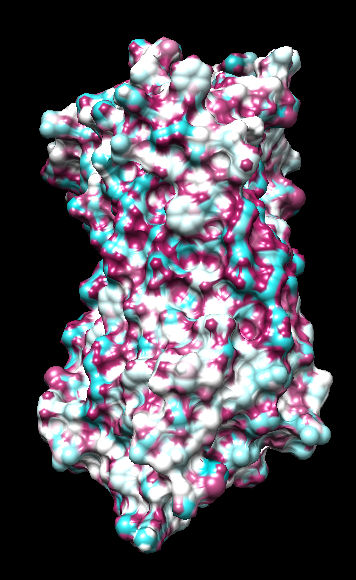
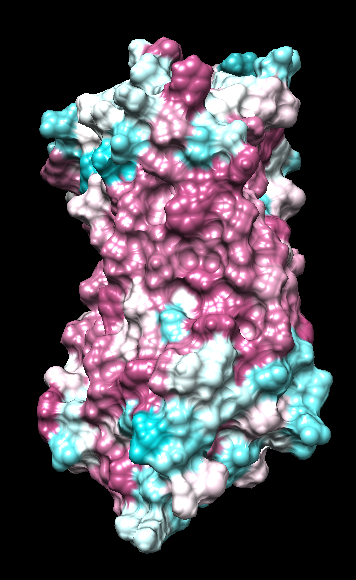
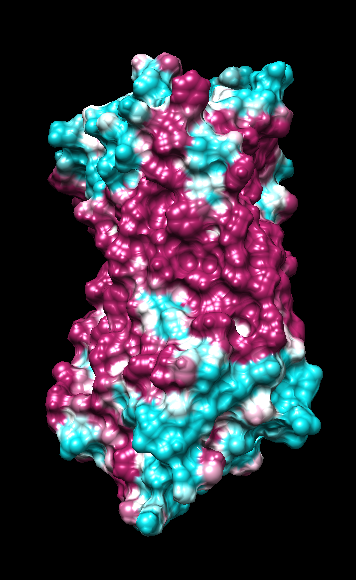

range -1.76, 0.76
("bfactor" in 1hzxAwithH_atomic.pdb)
range -0.17, 0.11
(Chimera calc residue avg "bfactor")
range -1, 1
("bfactor" in 1hzxAwithH_centers.pdb)
range -1.5, 1.83
coloring -1,0,1
(all others min,0,max)
Elaine Meng
Aug-Sep 2016; 2019 update below
MLP (or MHP for molecular hydrophobicity potential) is a construct that spreads atomic values out in 3D, analogous to an electrostatic potential calculated from atomic partial charges. It is not clearly defined, so various different functional forms are used, as well as different sets of atomic parameters. Most of the emphasis in papers about the atomic contributions is on using them to predict the logP (octanol-water partition coefficient) of entire small organic molecules. Coloring/display of “hydrophobicity potential” is a later application.
Qualitative conclusion from the limited set of comparisons below: for protein depiction, a simple amino acid lookup may be be better than or at least as good as than a potential, given the latter's complexity, computational demands, and dependence on reasonable atomic values. See for example the result of simply averaging PLATINUM atomic values over residues without using a distance-dependent equation (below). The atomic values used by pyMLP and thus mlp in ChimeraX may not be that good; however, for the comparisons here I did not explore the different functional forms and other adjustable parameters in mlp (details...). The possible advantage of an atom-type-based potential is that it could be used on arbitrary organic compounds, but this has not been implemented in ChimeraX yet. Ideally we'd have both:
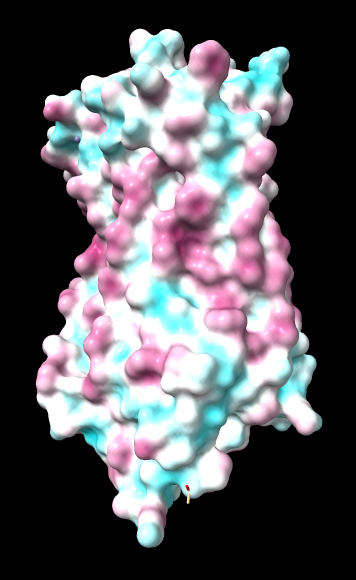
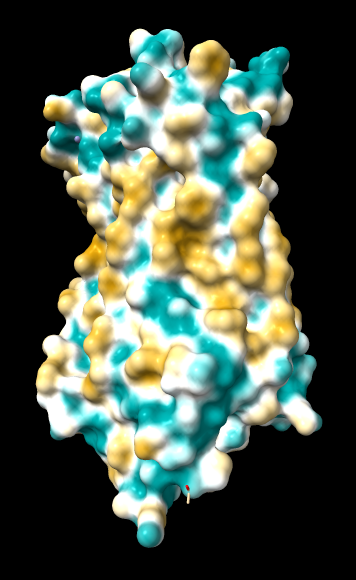
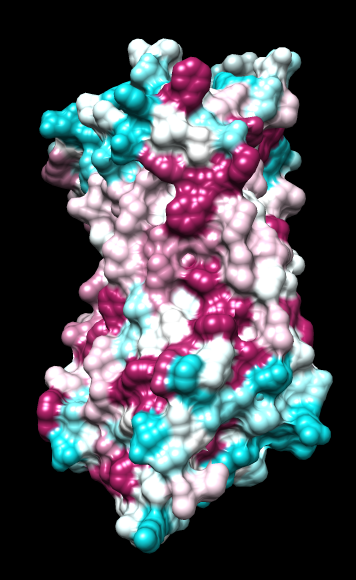
Aug 17, 2016. Trying the PLATINUM server http://model.nmr.ru/platinum/ on membrane protein rhodopsin, 1hzx chain A:
... compare to current Chimera and ChimeraX options:
 |
 |
 |
 |
 |
 |
 |
 |
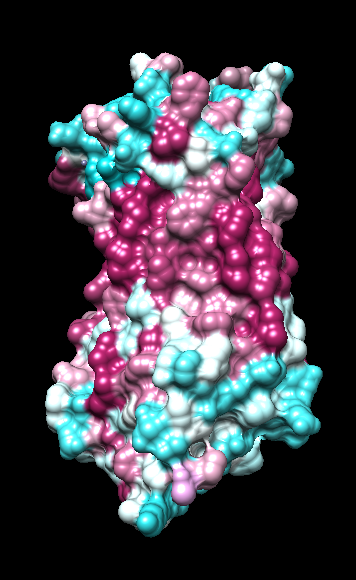
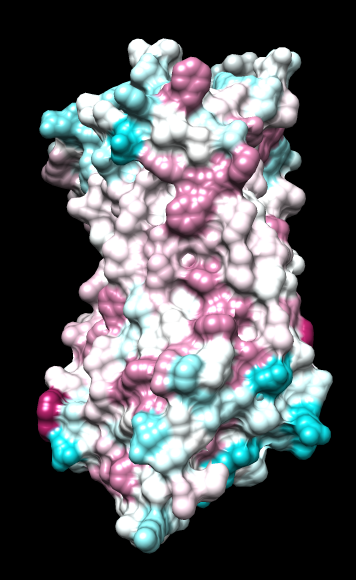
| ChimeraX mlp
default parms except color min,0,max min -45, max 25 |
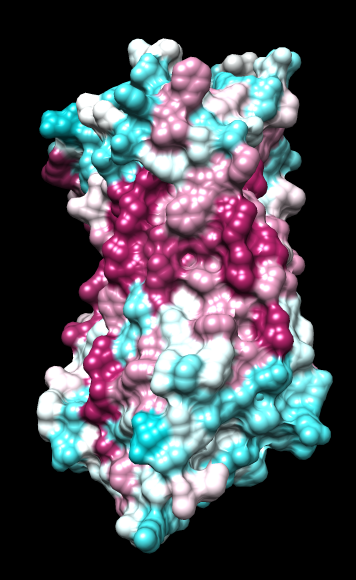
ChimeraX mlp
same except color -20,0,20 (default coloring range) |
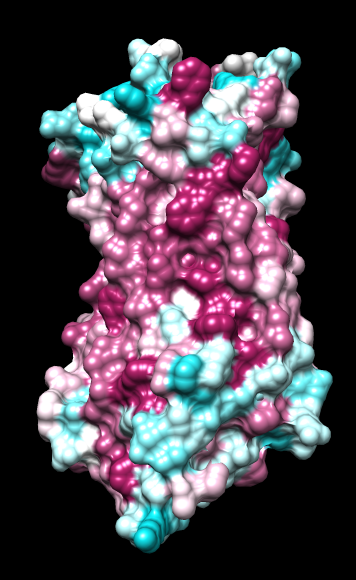
ChimeraX mlp
default everything |
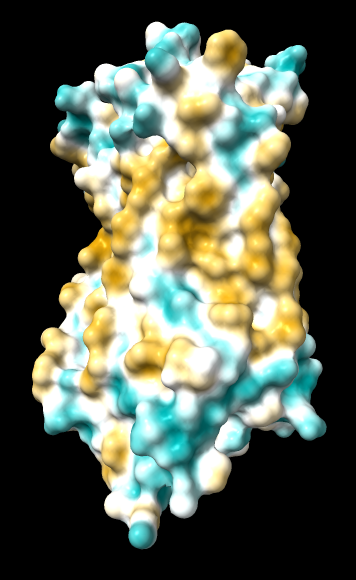
Chimera
kdHydrophobicity color min,0,max |
Chimera
wwHydrophobicity color min,0,max |
Chimera
wwHydrophobicity color -1,0,1 |
Chimera
hhHydrophobicity color min,0,max *negative is more hydrophobic in this scale* |
Chimera
mfHydrophobicity (suggested by Oliver Clarke, see: http://plato.cgl.ucsf.edu/pipermail/chimera-users/2016-July/012562.html) color min,0,max *negative is more hydrophobic in this scale* |
Files for/from the PLATINUM server (more details in their manual):
Distance function:
Due to some limitations of the original set of atomic lipophilicity values from pyMLP, most noticeably asymmetry and sign difference in the values for ASP/GLU carboxylate oxygens, I investigated using the atomic lipophilicity values in the Ghose paper instead. These were developed for ALOGP/CLOGP calculations and are used by the PLATINUM webserver.
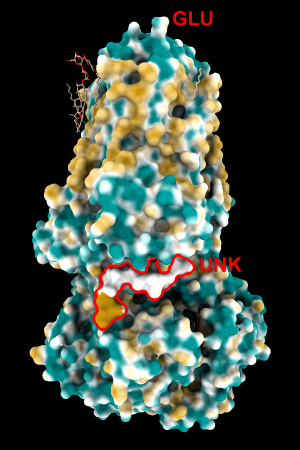
This paper lists very many atom types, much more subdivided than the ChimeraX atom types (for example, nine different types for just H bonded to C!). It's not feasible for us to encode the chemical rules to recognize all of these types currently, and we want to allow MLP coloring without requiring hydrogen addition. Thus, I manually created a lookup table with the same amino acids as the original pyMLP set in which the values for the (inferred) attached hydrogens were added to the respective heavy-atom values. I'll call this set of atomic parameters ghose-united. To the original set of residues, I added a few more types that could occur within a protein chain: MLE and the peptide-capping residues NH2, NME, and ACE, as well as UNK (backbone only, sometimes used for lower-resolution structures in which the amino acid type cannot be determined from the density).
I also made a ghose-united-shifted set incorporating the 0.03 shift per atom recommended in the Platinum manual, accounting for the 0-3 hydrogens that had been united with each heavy atom. These files can be tested by being substituted for the file mlp.py (keeping that name) in the ChimeraX download.
Numerical results, images, and conclusions as to what we should use are given below.
Assumptions and judgment calls:
I tried the ChimeraX mlp command with these different atomic-value sets on several proteins, including:
structure min, mean, max on surface (rounded) original ghose-united ghose-united-shifted 1hzxA -45, -5, 25 -27, 0, 24 -21, 4, 30 1a0sQ -48, -8, 25 -34, -5, 24 -28, -1, 28 3mhc -45, -9, 23 -31, -5, 23 -26, -2, 28 3w7fA -45, -7, 25 -26, -4, 25 -21, 0 30 1bxw -50, -9, 22 -28, -4, 23 (not tried) 6o2pA -48, -6, 26 -29, -2, 26 (not tried) 6o2pB
(has only UNK res, bb atoms)0,0,0 -24, -11, -2 (not tried)
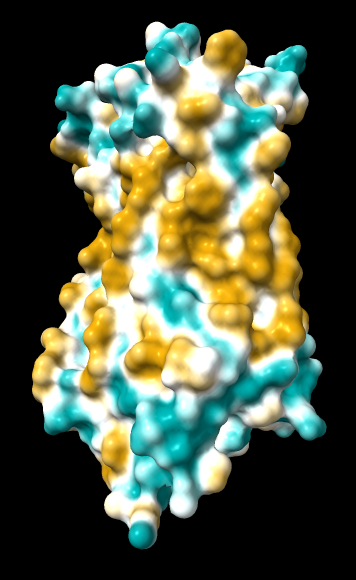
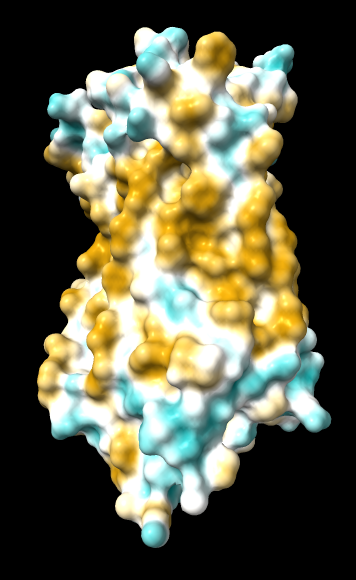
1hzx chain A, default coloring range (-20,20) except where noted otherwise:
| original | ghose-united | ghose-united (range –15,15) |
ghose-united-shifted |
 |
 |
 |
 |
|
ghose-united
(same as above except Asp/Glu highlighted) |
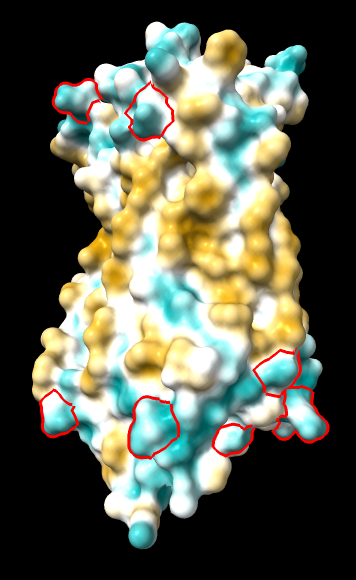 |
A few more of the protein examples, default coloring range (-20,20):
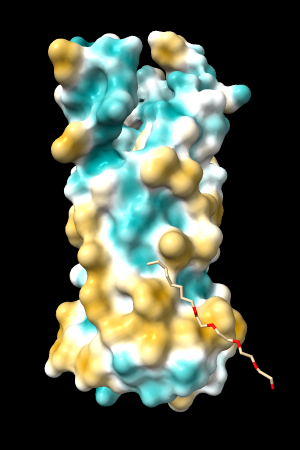
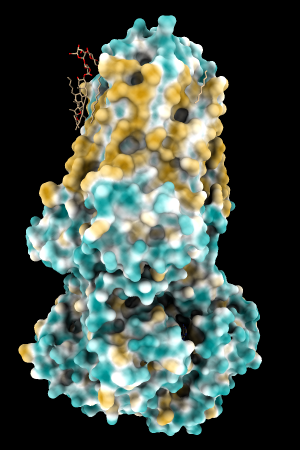
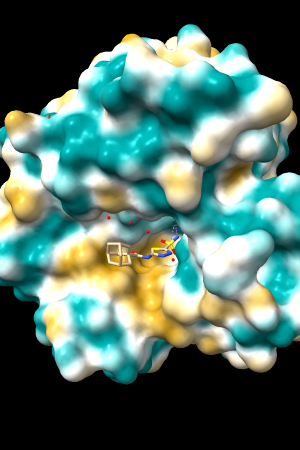
| 1bxw | 6o2p | 3mhc | |||
| original | ghose-united | original | ghose-united | original | ghose-united |
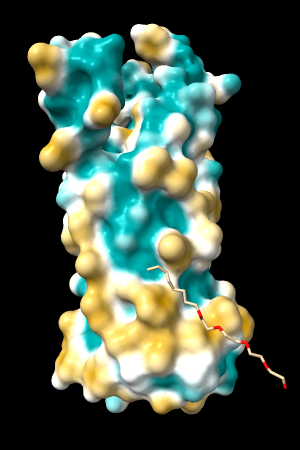 |
 |
 |
 |
 |
 |
Quite consistently on a variety of proteins, the original parameters give a minimum of negative 40-50, mean of negative 5-10, and max in the 20s, whereas the ghose-united parameters shift the minimum to negative 25-35 and the mean closer to zero, but give about the same maximum as the original parameters. Coloring over the same default range (-20,20) gives the same overall impression, with the same regions coming out as hydrophobic, and not surprisingly, a more reasonable result for Asp/Glu sidechains and UNK/backbone-only residues. I recommend using the ghose-united parameters as the new default and keeping the default coloring range the same. The additional ad hoc shift (ghose-united-shifted parameters) doesn't seem to add anything to the analysis. Anyone who prefers a more saturated coloring could use a more restricted coloring range (e.g. -15,15) but I think the -20,20 range provides a better sense of the range of values anyway. (A frequent problem in paper figures is the use of oversaturated coloring for electrostatic potential.)