
Match -> Align creates a sequence alignment from a structural superposition of proteins or nucleic acids in Chimera. Residue types are not used, only their spatial proximities. Iterations of refitting the structures using the sequence alignment and generating a new sequence alignment can be performed.
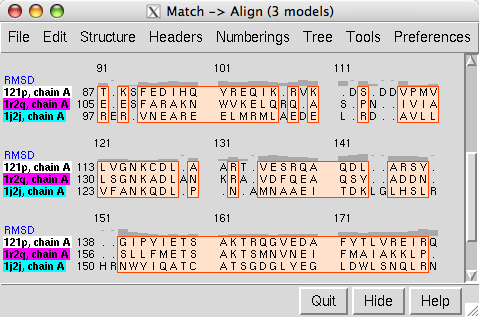
The output sequence alignment is automatically shown in Multalign Viewer, and RMSDs over the fully populated columns of the alignment are reported in the Reply Log.
 |
For an informal introduction, see the Superpositions and Alignments tutorial. See also: MatchMaker, Multalign Viewer, and
Tools for integrated sequence-structure analysis with UCSF Chimera. Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. BMC Bioinformatics. 2006 Jul 12;7:339.There are several ways to start Match -> Align, a tool in the Structure Comparison and Sequence categories.
Chains to be included in the sequence alignment should be chosen from the top section of the panel.
Iteration Parameters:
- Iterate alignment:
- at most [N] times (default 3) - refit and then generate a new sequence alignment N times (or fewer, if convergence is reached)
- until convergence - refit and then generate a new sequence alignment until the number of fully populated columns no longer increases
- Superimpose full columns:
- across entire alignment - refit the structures using all fully populated columns of the sequence alignment
- in stretches of at least [L] consecutive columns (default 3) - refit the structures using only the fully populated columns in consecutive stretches of L or more
- Reference chain for matching [chain] - which structure should remain fixed as the others are matched to it
Clicking Apply (or OK, which also dismisses the dialog) initiates the calculation.
The output sequence alignment is automatically shown in Multalign Viewer and can be saved to a file from that tool. The fully populated columns are highlighted as a region (colored boxes). Clicking the region will select the corresponding parts of the structures, in effect their common cores. The header named RMSD shows the spatial variation per column.
The number of fully populated columns in the alignment and the corresponding pairwise and overall RMSDs are reported in the Reply Log. All RMSDs are calculated using one atom per residue: CA in amino acids, C4' in nucleic acids. Structures are not refit using the final sequence alignment; rather, the existing superpositions are simply evaluated over the fully populated columns of that alignment.
Close dismisses the dialog without generating an alignment. Help opens this manual page in a browser window.
In most cases, a semi-heuristic algorithm is used. However, a modified Needleman-Wunsch procedure (dynamic programming) is used for the case of two chains and no allowance for circular permutation:
The score for aligning a pair of residues is: The gap penalty is zero, since for this application the spatial proximity should be more important than adjacency in sequence; that is, residues farther apart than the distance cutoff should not be aligned.This process determines the sequence alignment that best represents the structural alignment.