This tutorial describes superimposing structures "manually" (by explicitly specifying the atoms to be matched), saving positions and sessions, and creating publication-quality images.
The subjects are a set of glycoside hydrolases with shared structural features, as described in the papers:
T. Pons, D.G. Naumoff, C. Martinez-Fleites, and L. Hernandez, "Three Acidic Residues Are at the Active Site of a Beta-Propeller Architecture in Glycoside Hydrolase Families 32, 43, 62, and 68" Proteins 54:424 (2004).
F. Alberto, C. Bignon, G. Sulzenbacher, B. Henrissat, and M. Czjzek, "The Three-Dimensional Structure of Invertase (Beta-Fructosidase) from Thermatoga maritima Reveals a Bimodular Arrangement and an Evolutionary Relationship between Retaining and Inverting Glycosidases" J Biol Chem 279:18903 (2004).
| [images shown at 25%; click for full size] |

|

|
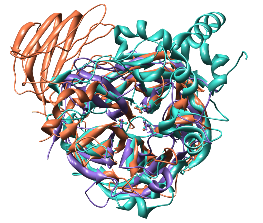
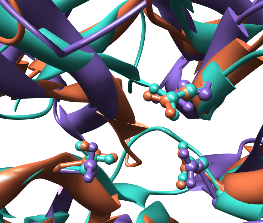
| Comparison of glycoside hydrolases from different families. Invertase (family 32) is coral, arabinanase A (family 43) is purple, and levansucrase (family 68) is turquoise. Despite differences in sequence, substrate, and catalytic mechanism, the enzymes have similar overall folds (top) and a set of highly conserved acidic residues in the active site (bottom). |
Glycoside hydrolases (GH) are a large, heterogeneous set of enzymes that catalyze the hydrolysis of certain bonds between sugars or between sugars and other moities. Based on sequence comparisons, these enzymes have been categorized into many families; see the CAZy web site for details.
This tutorial describes the creation of images to show structural similarities among certain members of GH families 32, 43, and 68. These enzymes were grouped into different families because their relationships were not clear from sequence information alone.
The approach described on this page is only one of many possibilities. Another is to generate static renderings with shadows.
On Windows/Mac, click the chimera icon. On UNIX, start Chimera from the system prompt:
unix: chimera
A basic Chimera window should appear after a few seconds; resize it as desired. Open the Command Line (choosing Tools... General Controls... Command Line is one way).
If you have internet connectivity, structures can be obtained directly from the Protein Data Bank. Choose File... Fetch by ID from the Chimera menu. In the resulting dialog, check the PDB ID option (if it is not already checked) and the option to Keep dialog up after Fetch. Fetch the PDB structures 1uyp, 1gyd, and 1oyg, in that order, and then Close the dialog. If you do not have internet connectivity, you can download the files 1uyp.pdb, 1gyd.pdb, and 1oyg.pdb into your working directory and then open them in that order as local files (with File... Open). (Or instead of opening these structures, one could just open the Chimera session that is saved below, but that would not be very instructive as to how the session was generated.)
Move and scale the structures with the mouse in the graphics window (and optionally, the Side View) as desired throughout the tutorial.
Some salient features of the structures:
| PDB ID | enzyme | family | chains | conserved residues | ||
|---|---|---|---|---|---|---|
| 1uyp | invertase, T. maritima |
GH32 | A-F | Asp17 | Asp138 | Glu190 |
| 1gyd | arabinanase A, C. japonicus |
GH43 | B | Asp38 | Asp158 | Glu221 |
| 1oyg | levansucrase, B. subtilis |
GH68 | A | Asp86 | Asp247 | Glu342 |
First, simplify the situation by deleting unwanted (for our purposes) portions of the structures such as extra chains and solvent, and then displaying just the alpha-carbon traces.
Command: delete #0:.b-fBecause the sequences are quite dissimilar and a set of structurally equivalent residues are known, the most efficient way to superimpose the structures is by specifying which atoms/residues should be used for the least-squares fit:
Command: delete solvent
Command: chain @ca
Command: linewidth 3
Command: match #1:38.b:158.b:221.b@n@ca@c@o #0:17.a:138.a:190.a@n@ca@c@oHere, we used the peptide backbone atoms of the conserved residues. Another possibility would have been to use all atoms of these residues, as the residue types are the same across the different structures (note that match requires equal numbers of atoms from the two models being matched). Model 0 (invertase) was used as the reference structure for both pairwise matches. The number of points used for fitting and the resultant RMSD values are reported in the status line (transiently) and the Reply Log.
Command: match #2:86.a:247.a:342.a@n@ca@c@o #0:17.a:138.a:190.a@n@ca@c@o
Show the structures as ribbons, with only the side chains of the conserved residues shown in ball-and-stick form:
Command: ~dispEven though all atoms of the residues are selected, by default the backbone atoms will not be displayed at the same time as ribbon (this can be controlled with the command ribbackbone).
Command: ribbon
Command: sel #0:17.a:138.a:190.a #1:38.b:158.b:221.b #2:86.a:247.a:342.a
Command: disp sel
Command: repr bs selThe entire protein chains are being shown. You can undisplay the ribbon for a portion of a chain:
Command: ~sel
Command: ~ribbon #0:296-end.aThis was not done for the examples, however.
The background can be any color, but white is often good for publication.
Command: set bg_color whiteOther ways to change the background color are with the menu (Actions... Color) and in the Background preferences. Further, if system hardware permits, starting Chimera with the --bgopacity option enables background transparency. Images saved with a transparent background are easier to composite with different backgrounds in image-editing applications.
When the background is white, the default type of depth cueing (shading from front to back with increasing amounts of black) looks strange. Use the Effects tool (Tools... Viewing Controls... Effects) to turn off depth cueing (uncheck the checkbox). Another reasonable approach is to make the depth cueing color white instead of black, then adjust the amount of white shading by moving the clipping planes in the Side View. Close the Effects tool.
Straight edges may appear jagged, but but supersampling will correct these within saved images.
Color the three models three distinguishable colors.
Command: sel #0Repeat for models 1 and 2. For the example images, the colors named coral, medium purple, and turquoise were chosen for models 0, 1, and 2, respectively. Clear the selection (Select... Clear Selection is one way).
Actions... Color... all colors [pick a color]
Open Shininess Control (Tools... Viewing Controls... Shininess Control). Set the Shininess to 128 and the Brightness to 5, then Close the tool.
Additional effects can be achieved by adjusting the Lighting, although this was not done for the examples.
The default ribbon style is flat; the edged and round styles are more three-dimensional.
Command: ribrepr roundSometimes the ribbons obscure the view of important side chains or other moieties. You can change the ribbon scaling (height/width) using the Ribbon Style Editor. Open this tool (Tools... Depiction... Ribbon Style Editor) and if necessary, click the tab to bring the Scaling panel to the front. For the example images, all width and height settings were reduced by 20% from the Chimera default values. Scalings can be named and saved for later use (the information is stored in the preferences file).
The smoothness of curved surfaces in the stick, ball-and-stick, sphere, and ribbon molecular representations can be increased by raising subdivision quality in the Effects tool (Tools... Viewing Controls... Effects). Higher values increase smoothness but may slow performance. Values of 5.0-20.0 are reasonable for saved images, while lower values suffice for interactive use. 5.0 was used for the examples.
Similarly, the smoothness of molecular surfaces (not included in this example) can be increased by increasing the vertex density in the surface attributes panel.
Straight edges may appear as tiny zigzags, especially against a white background. Supersampling gets rid of such jagged edges within saved images by making an initial image larger than that requested and then sampling it down to the final size. The level of supersampling can be controlled in the Image Setup preferences. The default value of 3x3, used for the examples, is generally sufficient for publication.
Resize the Chimera window to the desired aspect ratio, and then find view(s) suitable for the images. For the example images, a position was found that showed the overall structures well, and when scaled up (without further rotation) showed the conserved side chains well. The zoomed-out position was named overall, the scaled-up position was named closeup, and a session file named images.py was saved:
Command: savepos overall(To browse to a save location, use File... Save Session As instead of save.) In this example, the session was saved in the overall position, but it could have been saved in another position (including one without a name) without any loss of information. It is generally prudent to save sessions for publication images, as this decreases the labor necessary if figures have to be redone. Often minor adjustments such as changing a color or displaying a different set of side chains will be required.
Command: scale 4
Command: savepos closeup
Command: reset overall
Command: save images
Later, the session can be restarted by opening images.py. If the models are moved around, the saved positions can be regenerated, for example:
Command: reset closeupIt should be noted that some things are not saved in a session; additional operations may be required to regenerate images exactly.
Before an image is saved, a particular position may need to be restored, for example:
Command: reset overallThe overall and closeup positions were used for the examples.
Choosing File... Save Image brings up the Save Image panel. Editing the value for one dimension and then clicking in the field for the other dimension adjusts the latter automatically when Maintain current aspect ratio is on. The dimensions can be specified directly in pixels, or in units of length. If units of length, the number of pixels per unit can be controlled in the Image Setup preferences.
For example, to make an image for a single-column width of 85 mm (3.346 in) with a resolution of at least 300 dpi, possible approaches are to:
The example images were saved in TIFF format; other possibilities are PNG, JPEG, PS (PostScript), and EPS (encapsulated PostScript).
Labeling. The standard labels in Chimera (those generated with the Label section of the Actions menu or the commands label and rlabel) have controllable content, font, size, and color, but there is little control over position. However, labels appropriate for publication/presentation images can be generated with the 2D Labels tool or its command equivalent, 2dlabels. These labels have controllable content, size, and color, and can be placed anywhere in the x,y coordinate system of the graphics window.
Color space. For some publications, images must be in the CMYK color space. Chimera currently saves images in only the RGB color space, so a separate application such as Adobe Photoshop® must be used to switch between the two.
Stereo. To generate stereo images, change the camera mode setting in the Camera tool before saving an image file. Due to current problems with the cross-eye stereo and wall-eye stereo options, the best approach is to save separate images in the stereo left eye and stereo right eye modes and assemble them in the desired order to give a cross-eye or wall-eye stereo image. (Camera modes can also be switched with the stereo command.)