

 home
overview
research
resources
outreach & training
outreach & training
visitors center
visitors center
search
search
home
overview
research
resources
outreach & training
outreach & training
visitors center
visitors center
search
search


 home
overview
research
resources
outreach & training
outreach & training
visitors center
visitors center
search
search
home
overview
research
resources
outreach & training
outreach & training
visitors center
visitors center
search
search

Tom Goddard¹, Matthew Baker², Wen Jiang², Wah Chiu²
¹
Computer Graphics Laboratory,
University of California, San Francisco
²
National Center for Macromolecular Imaging,
Baylor College of Medicine
Wah Chiu's lab at the Baylor College of Medicine focuses on the use of electron crystallography and cryomicroscopy to determine the three-dimensional structures of macromolecular assemblies at atomic resolution.
The Computer Graphics Lab is developing software to interactively explore microscopy data and molecular structures.
All images on this page ©2004 The Regents, University of California; all rights reserved.

|
Helixhunter ExampleChimera is used to examine a density map and the locations of helices predicted by helixhunter. The example looks at a Bluetongue virus capsid protein density map simulated at 8A resolution from crystal structure 1BVP. This is in the range of resolutions achievable by electron cryomicroscopy reconstructions based on several thousand particles. |

|

|
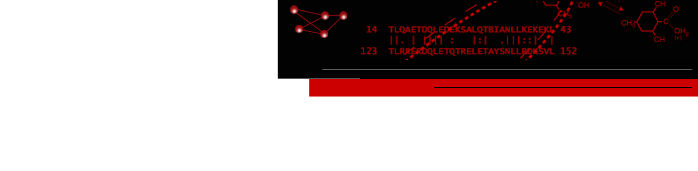
Tracing Connections between HelicesThe helices found in a density map can be matched with segments of the amino acid sequence that are predicted to fold into helices. The correspondence can be inferred by tracing the connections between helices in the density map to give a full picture of the protein backbone. Manual tracing can be done in Chimera by placing markers which are linked to form a chain. |
Laboratory Overview | Research | Outreach & Training | Available Resources | Visitors Center | Search