

 |
 |
Screen shots were taken from either Safari or Firefox, depending upon which produced the clearer, more easily read image.
| Table of Contents: |
| #1 | What is the scope of information available at NCBI on cystic fibrosis in humans? |
| Hints: | Do a All Databases search at NCBI (http://www.ncbi.nlm.nih.gov). Then repeat the search narrowing the returned hits to human. |
| Answer: Lots of data to be explored. |
#1 step by step instructions
| 1. | Go to http://www.ncbi.nlm.nih.gov, |

| enter cystic fibrosis in the for box |
| and click Go. | |
| 2. | The returned Entrez page is organized with literature matches in the top box, sequence based information in the middle one and NLM's resources in the bottom box. Items of possible interest are denoted by numbers in a white box next to a topic title. |
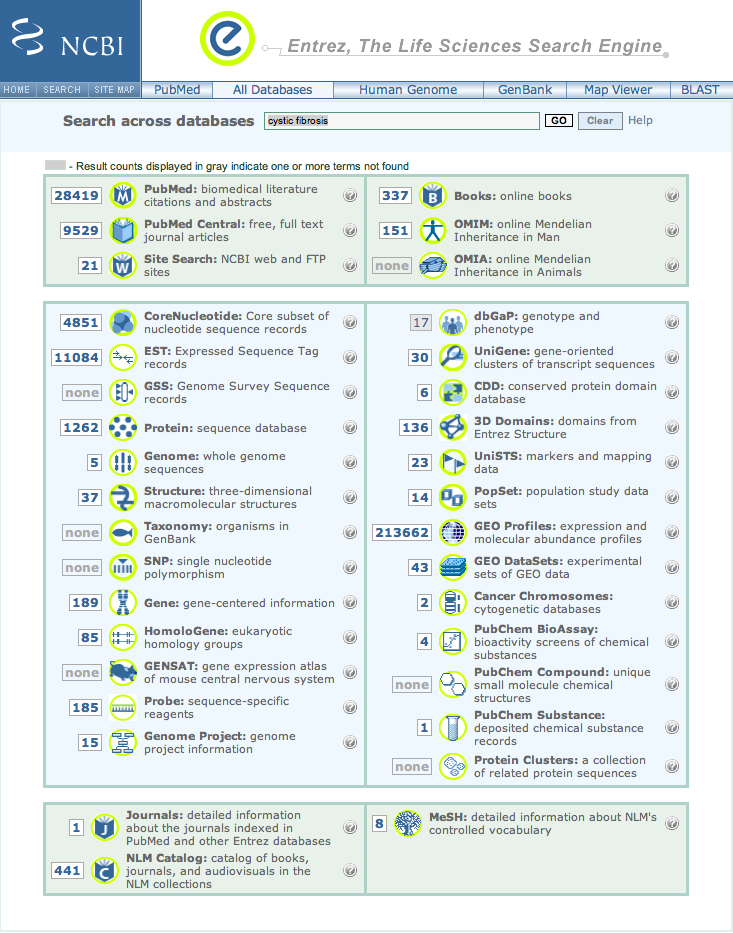
There are 151 OMIM entries [catalog of human genes and genetic disorders].
OMIM background information with links off to help and frequently asked questions
The sequence based information section contains data on cystic fibrosis from all species.
Some of these topics are sensitive to the addition of species information to the search query. OMIM and MeSH are examples of this.
| 3. | Return to the previous window. To restrict the sequence data to that from humans, by adding homo sapiens to the current cystic fibrosis in the Search across databases box |

| and click Go. |
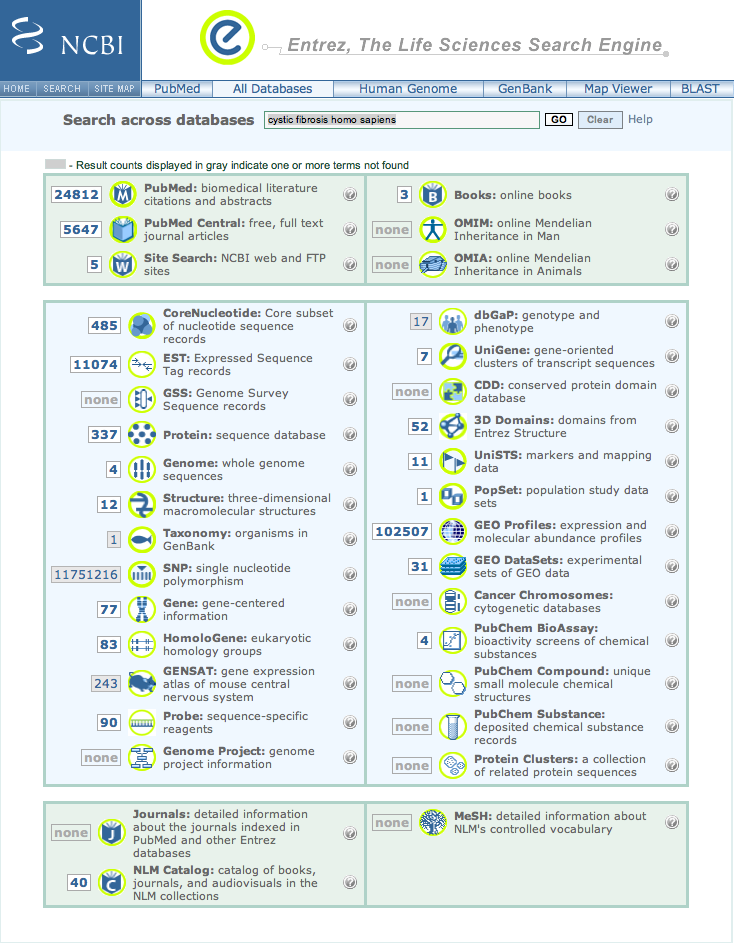
| 4. | The number of matches for nucleotide, protein, gene topics have decreased, but, there still are a very large number of items to sift through. |
| #2 | Besides the cystic fibrosis transmembrane conductance regulator gene (CFTR), what other genes are associated with cystic fibrosis in humans and what is their relationship to the disease? |
| Hints: | Perform an Entrez Gene search (http://www.ncbi.nlm.nih.gov) to find the other genes and their function or relationship to the disease. |
| Possible answers: |
| S100A8 | - | cystic fibrosis antigen |
| TGFB1 | - | mutations modify severity of pulmonary disease in cystic fibrosis patients |
| - | protein expression correlates with portal tracts showing histological abnormalities associated with cystic fibrosis liver disease | |
| GOPC | - | CFTR binding |
| ADRB2 | - | 2002 polymorphisms contribute to clinical severity and disease progressionin cystic fibrosis 2005 - transfected beta3 not beta2-adrenergic receptors regulates CFTR activity via new pathway |
| SLC9A3R1 | - | 6/2007 plays a role in the turnover of CFTR at the cell surface |
| - | 5/2007 modulation of the expression of CFTR protein partners, like NHE-RF1, can rescue sequence-deleted CFTR activity. | |
| ABCB1 | - | study to see how the common cystic fibrosis mutation might disturb transmembrane segments of the protein using ABCB1 as a model ABCB1 expression increases ATP release in respiratory cystic fibrosis cells potential clinical benefits discussed |
#2 step by step instructions
| 1. | Go to http://www.ncbi.nlm.nih.gov, |

| change the Search option from All Databases to Gene using the pull down menu, |

| enter homo sapiens cystic fibrosis in the for box |
| and click Go. |
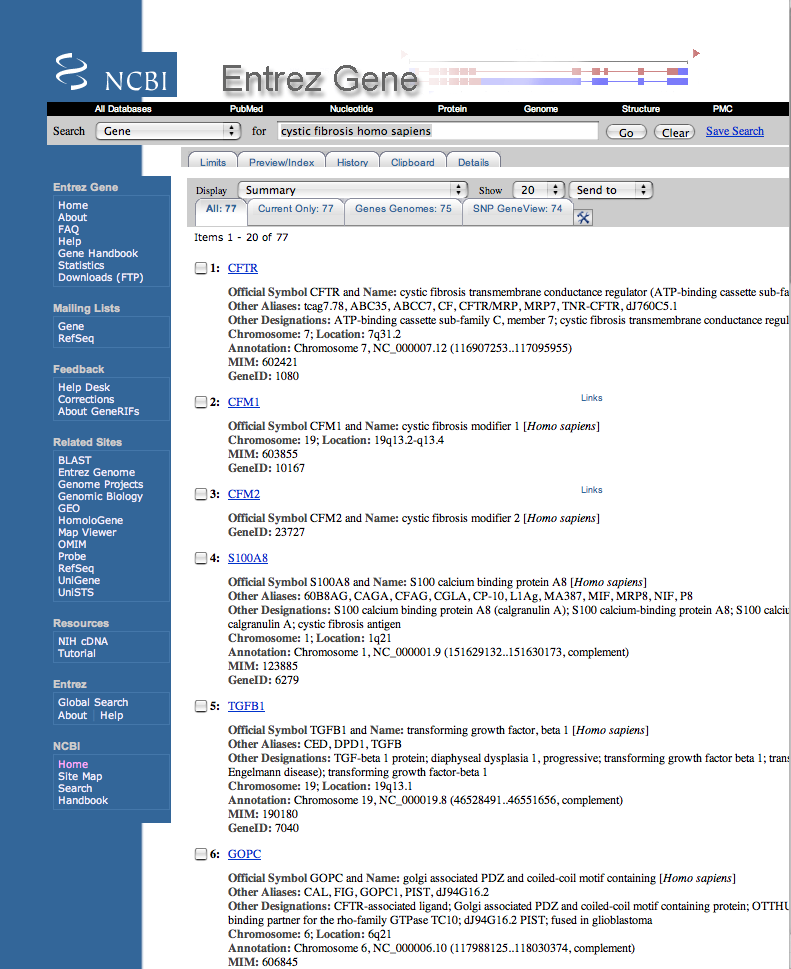
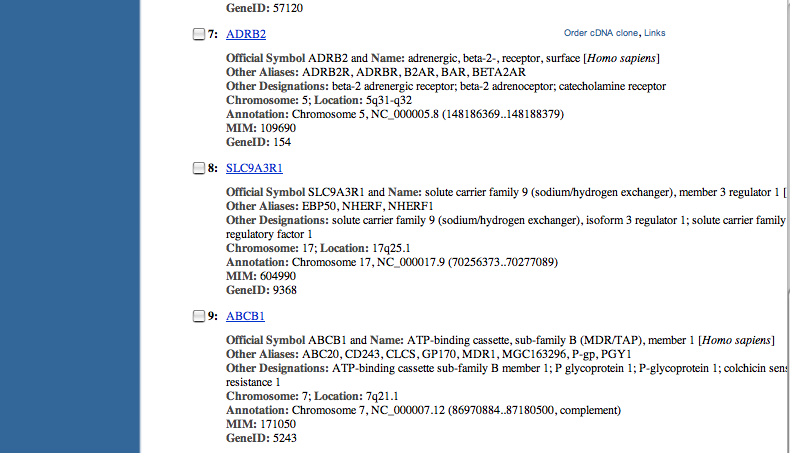
| Click on at least five diverse hits below the CFTR gene, finding out their relationship to cystic fibrosis. Ignore any gene that doesn't have a NCBI Reference Sequences (RefSeq) section. | |
| Here is what one of the results pages looks like. |
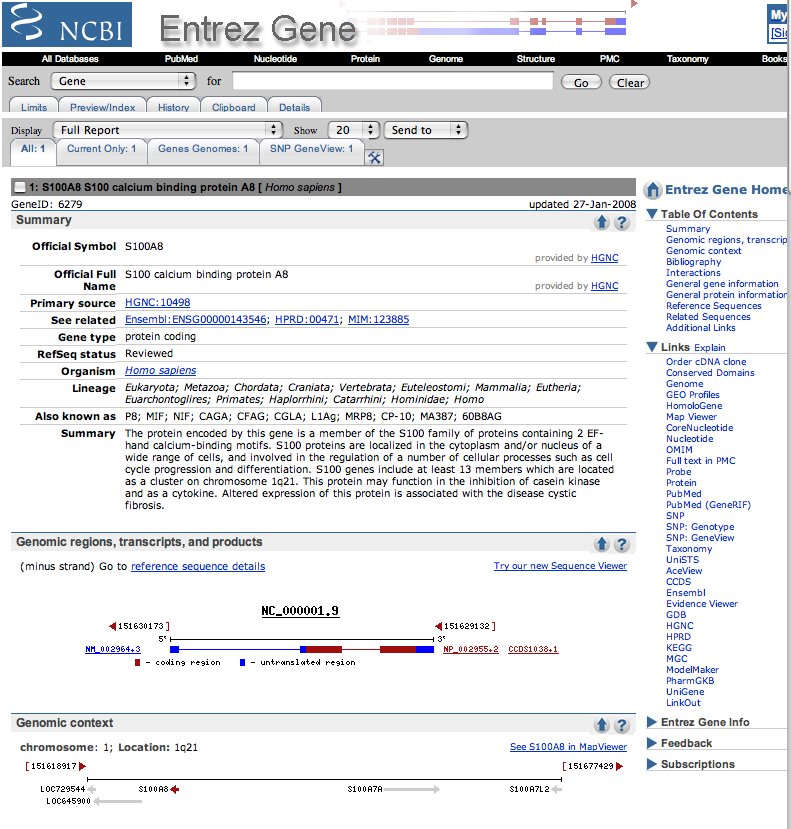
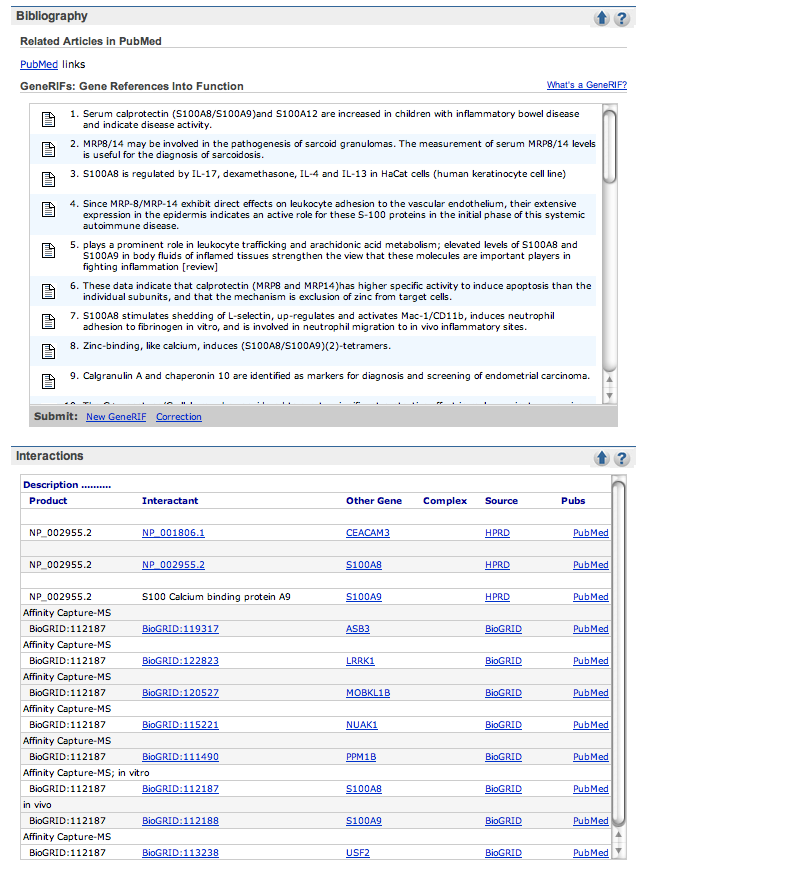
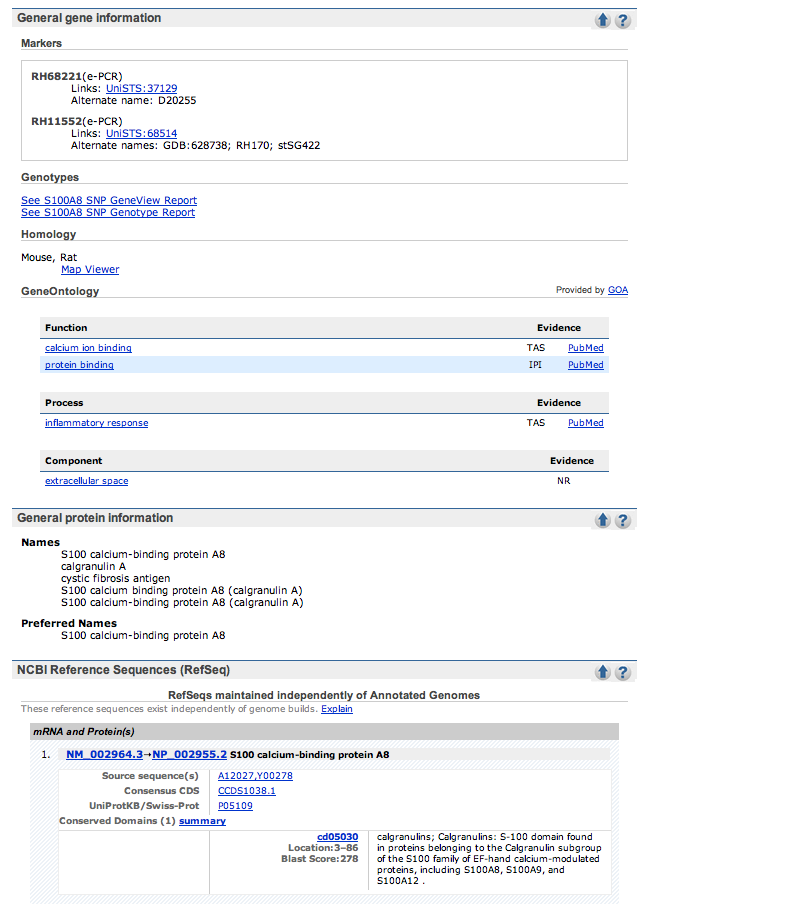
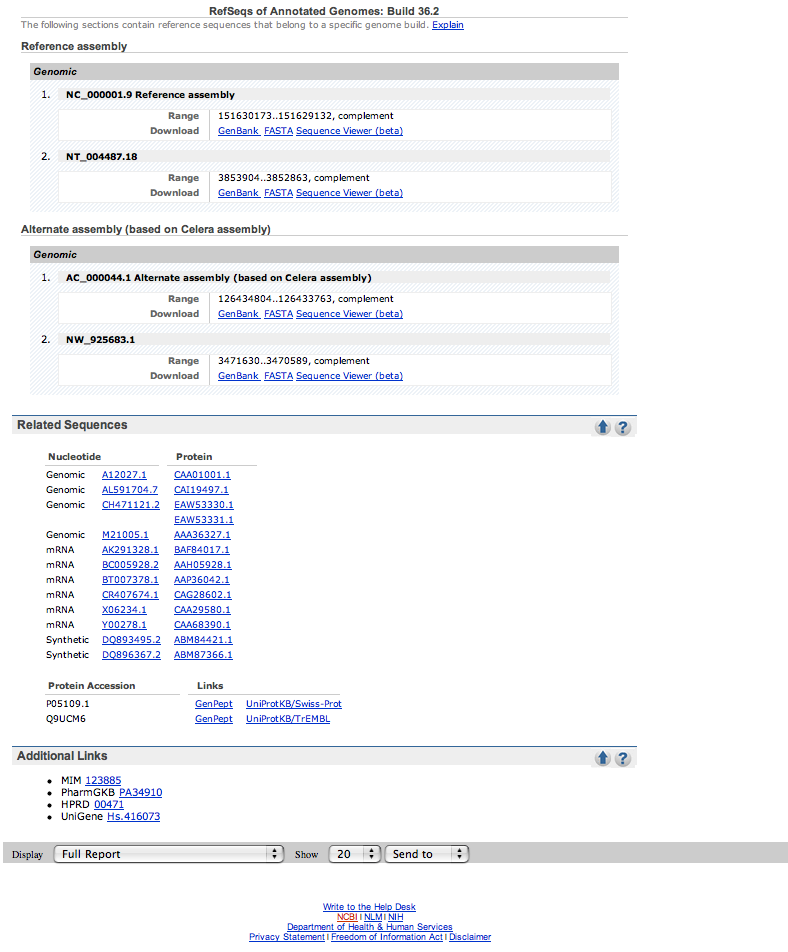
| Check out the Gene References into Function section of the Bibliography part of the page, if it exists. No hits, click on the PUBMED links in this section and scan through the titles for mention of cystic fibrosis. | |
| The given example (S100A8) had 4 pages of references and the cystic fibrosis ones were on the last page. | |
| If no papers are listed with cystic fibrosis in the description, check out the OMIM link on the side of the page or the links in the GeneOntology section. |
| Possible answers: |
| CFM1 | - | no RefSeq data (ignored) |
| CFM2 | - | no RefSeq data (ignored) |
| S100A8 | - | cystic fibrosis antigen |
| TGFB1 | - | mutations modify severity of pulmonary disease in cystic fibrosis patients |
| TGFB1 | - | protein expression correlates with portal tracts showing histological abnormalities associated with cystic fibrosis liver disease |
| GOPC | - | CFTR binding |
| ADRB2 | - | 2002 polymorphisms contribute to clinical severity and disease progressionin cystic fibrosis 2005 - transfected beta3 not beta2-adrenergic receptors regulates CFTR activity via new pathway |
| SLC9A3R1 | - | 6/2007 results indicate that NHERF1 plays a role in the turnover of CFTR at the cell surface, and that rDeltaF508 CFTR at the cell surface remains highly susceptible to degradation |
| SLC9A3R1 | - | 5/2007 modulation of the expression of CFTR (cystic fibrosis transmembrane conductance regulator) protein partners, like NHE-RF1, can rescue sequence-deleted CFTR activity |
| ABCB1 | - | study to see how the common cystic fibrosis mutation might disturb transmembrane segments of the protein using ABCB1 as a model ABCB1 expression increases ATP release in respiratory cystic fibrosis cells potential clinical benefits discussed |
| Note there is another database that is relevant for getting clinical information, Online Mendelian
Inheritance in Man (OMIM or MIM). Althought OMIM can not be searched via an actual sequence, it does allow searching
by gene symbol, chromosome location, keywords or other features. |
| Notice that gene names can change over time. SLC9A3R1 used to be called EBP50, NHERF, NHERF1 or NHE-RF1. |
| Hints: | Do a Gene search at NCBI (http://www.ncbi.nlm.nih.gov), record the codes. Compare the formats of the mRNA and protein sequences. Run a BLAST search, (http://blast.ncbi.nlm.nih.gov). |
| FASTA format is very concise, limited to the actual sequence and an identification line that starts
with a > symbol. The default format is very verbose, giving all sorts of reference details about the sequence
and a version of the sequence that is more easily read by the user. |
| BLAST searching allows for different types of data entry including the use of accession codes (such as a RefSeq accession code). |
| ADBR2 contains the 7tm_1 conserved domain signature which is highly conserved across species. |
#3 step by step instructions
| 1. | Go to http://www.ncbi.nlm.nih.gov, |

| change the All Databases Search option to Gene using the pull down menu, |

| enter homo sapiens nocturnal asthma in the for box |

| and click Go. | |
| 2. | Check the resulting hits to insure that the summary information on the gene mentions that various types of changes in this gene are associated with the disease. |
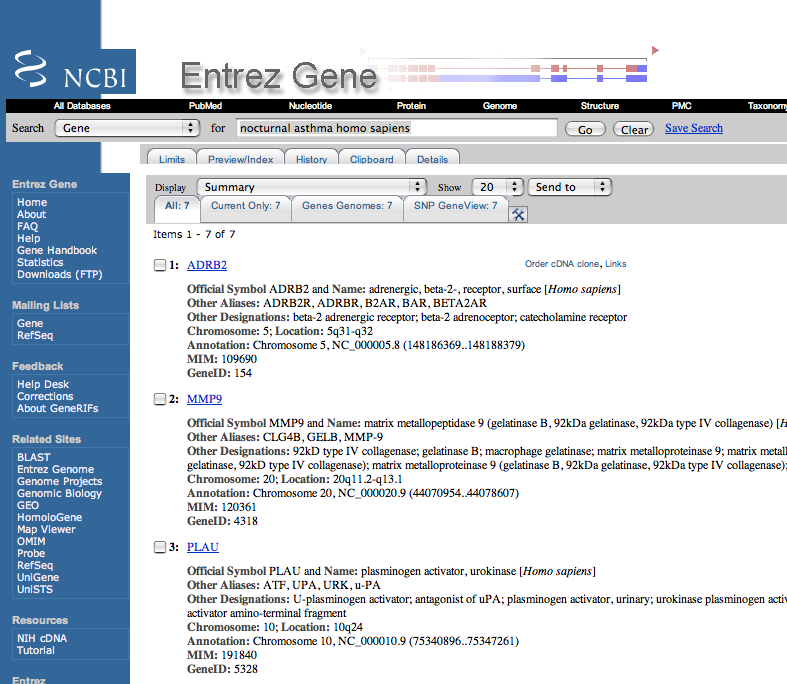
Here are the summary sections of the top three hits.
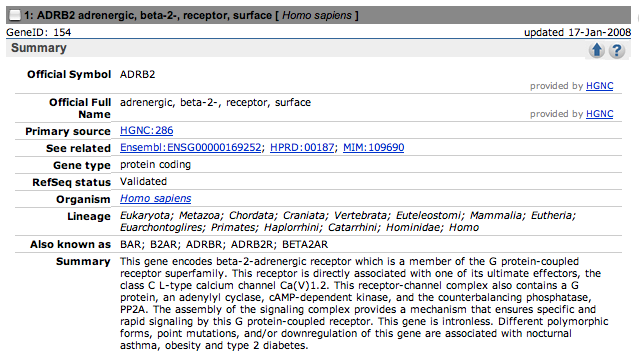

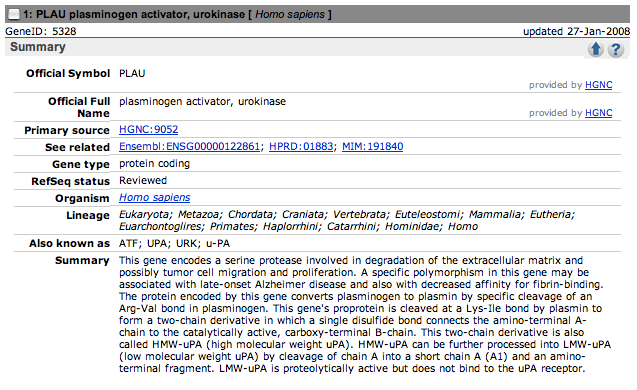
Only the first one contains a reference to nocturnal asthma.
| 3. | Scroll down the page to the NCBI Reference Sequences (RefSeq) section. Record the mRNA sequence and Product (protein) codes: |
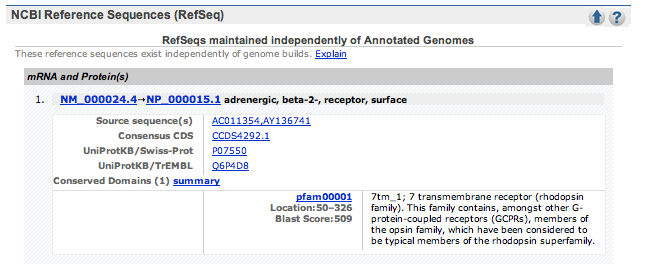
The required information:
| 4. | Click on the mRNA code to see the data on the actual mRNA sequence data. Scroll down the page taking in the
format of the information presented. |
| 5. | Scroll back to the top of the page and change the Display option from GenBank to FASTA. The format automatically changes. Note the difference. FASTA format is the sequence format required by many database searching programs.
|
| 6. | Click back to the Entrez Gene page and repeat this process with the protein code. |
| 7. | After noting the difference, click on the NCBI logo at the top of the page. |
| From the blue navigation bar on the main NCBI page, |

| click on BLAST. |
| 8. | From the main BLAST page, click on protein blastp in the Basic BLAST section. |
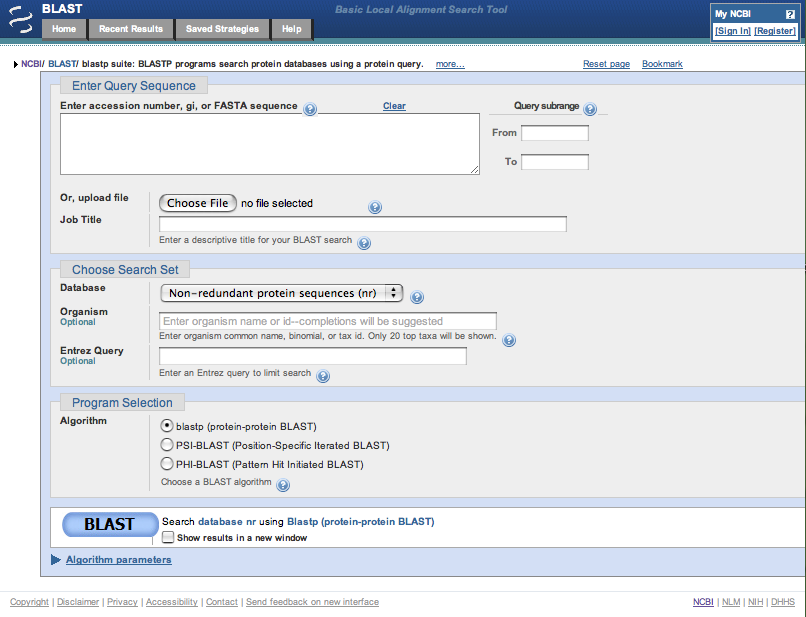
| In the blastp suite page, click on the ? icon at the top of the box in the Enter Query Sequence section to find out about what sort of inputs this form accepts. Clicking the more... link provides additional details. |
Clicking on the + symbol at the beginning of the green highlighted line produces the full version of the page.
After reading the presented information, click on the "?" icon again to close the information block and then enter the protein code
into the Enter Query Sequence box. Once this is done, information appears in the Job Title box.
In the Choose Search Set section of the page,and start to enter
the term Vertebrata into the Organism box.
As the term is entered, matching Entrez terms start to
appear. When enough of the word is entered to find the desired term, select this term from the list.
Clicking the BLAST button at the bottom of the page starts the search.

If results are to be displayed in a new window, click on the "Show results
in a new window" box prior to clicking the BLAST button.
Protein searches gets around the problem of multiple codons coding for the same amino acid that impacts
nucleotide searches. However, depending in the information sought, this is not always possible.
9.
It may take a few seconds for the search to be completed. While waiting, click on the 7tm_1 in the image to find
out about the conserved domain that was found in the sequence.
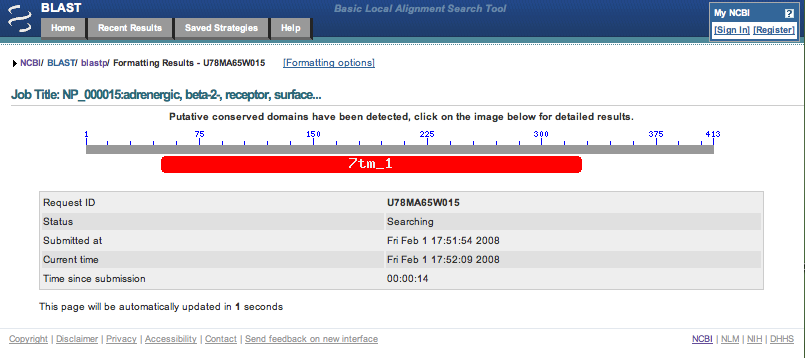
Initial Conserved Domains page.
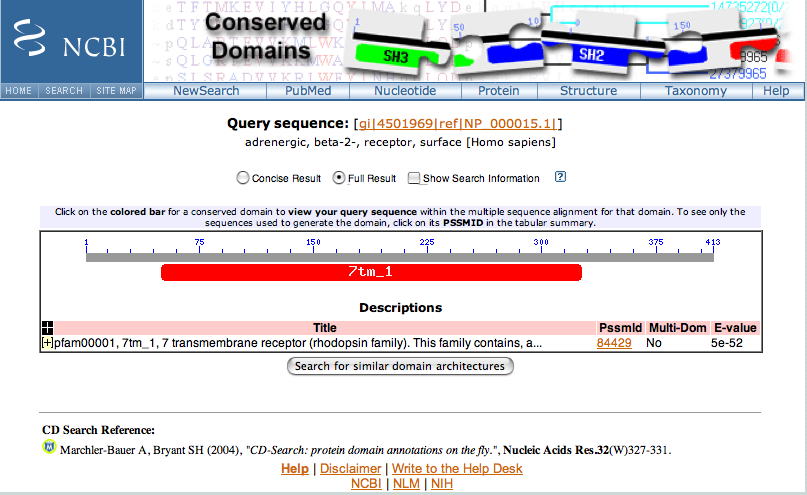
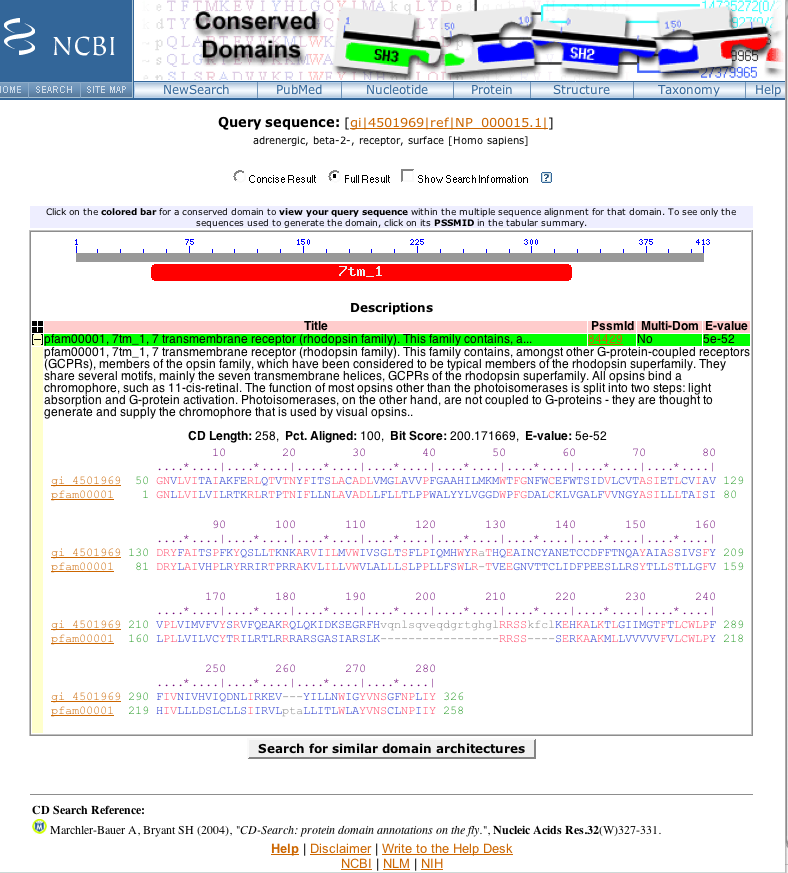
7tm_1 indicates that the protein being search with contains the transmembrane receptor signature of the
rhodopsin family of transmembrane proteins.
This signature is located in residues 50 to 326 of the sequence.
Close the popup window.
Note the length of the query sequence, this may be given on the Query line, the Job Title line or in the conserved domain image.
413 letters
Wait for the results page to appear.
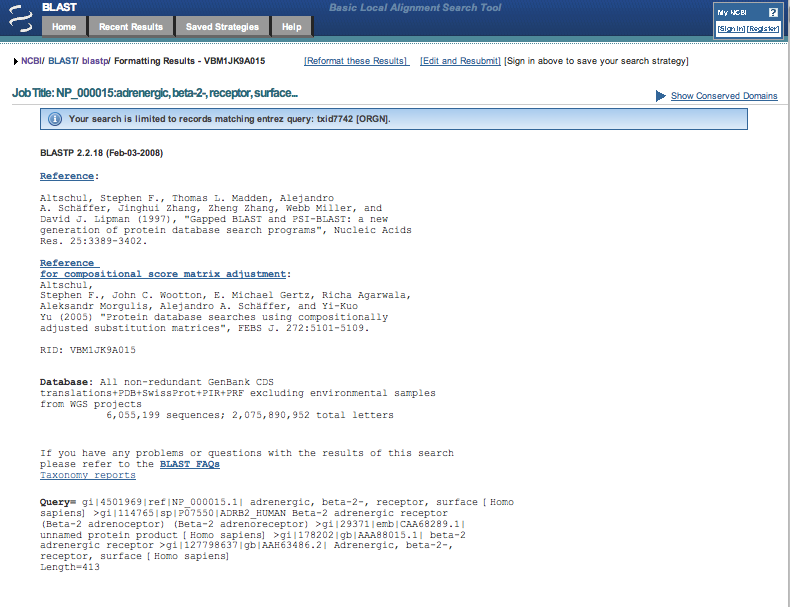
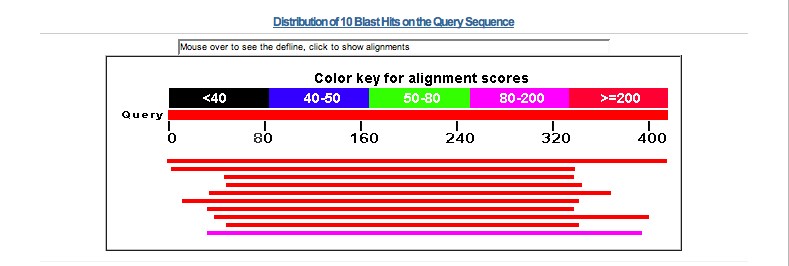
Here is a screen shot of the top part of the Blast results page.
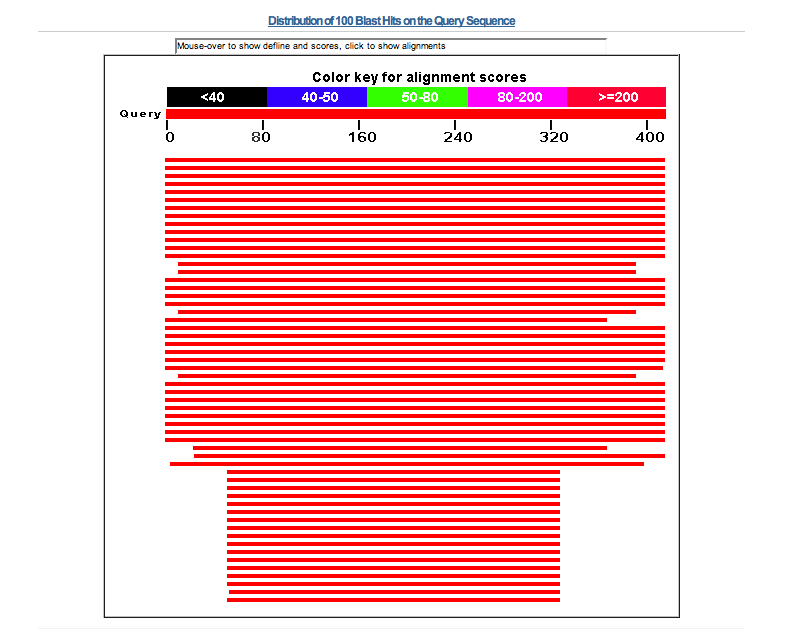
10.
Scroll down the results page past the image with its colored horizontal bars to the
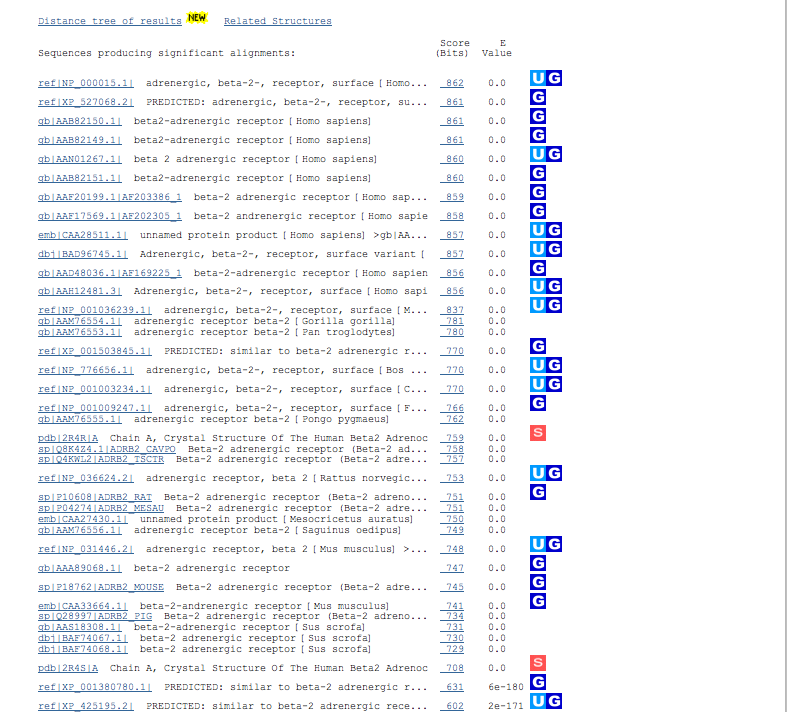
Sequences producing significant alignments section.
Scores are based on the length of the query sequence and the size of the database. Short sequences
will never produce great scores. To get a E value of 0.0 requires a match of at least 330 characters.
A very long sequence could easily have a match this long and still not have a match that covers a
significant portion of the query sequence. Always look at the resulting alignment. The mathematics
of the process can sometimes result in the strange ordering of hits.
A hit line gives the database the hit is from, its accession code, a description of the sequence from the database, its Bits score and finally
the E value. Hits in the list are ordered their E value, then their Bits score which reflects the
length of their actual match. Enough of the description may be given to see what species the match
is from.
Clicking on the link given on the left side of a hit line goes off to the actual sequence information.
Clicking on the right side link moves down to the alignment data for that hit.
Notice that there are over 30 hits with an E value of 0.0 at the top of this list and that the protein
code entered is at the top of the list. There are about 130 hits in the list which mention
ADBR2, beta-2 adrenergic receptor or variations thereof before sequence description changes
to something else. The first 12 hits are all from man with from 0 to 2 mismatches in the alignment.
NCBI used to make an effort to remove redundant sequences, but the size of the database increased to
such an extent that it was no longer possible to do this quickly enough so that it wouldn't impact
the processing of new data.
When an accession code begins with XP_, it means that the data is the results of an automated
analysis process. This situation usually occurs when a genome sequencing project is first being analyzed.
These sequences have not been checked for accuracy and can be much longer or shorter than their homologs
from more mature genome studies. These sequences usually have their description start with PREDICTED:.
The letter inclosed in colored boxes to the right of the hit line indicate that there is additional information
available about that matching sequence elsewhere. A boxed U means that there is Unigene data. A boxed G indicates
that there is Entrez Gene data. The boxed S means that there is structural data.
Check out some of the hits beyond the 0.0 E values and determine where the match is actually taking place within
the query sequence.
PREDICTED: similar to beta-2 adrenergic receptor [Monodelphis domestica] Length=404 (opossum)
results alignment section
PREDICTED: similar to beta-2 adrenergic receptor, [Gallus gallus] Length=397 (chicken)
12 - 396
results alignment section
beta-2 adrenergic receptor [Homo sapiens] Length=275 (man)
5 - 396
results alignment section
beta-2 adrenergic receptor [Macaca mulatta] Length=275 (rhesus monkey)
52- 326
results alignment section
beta-2 adrenergic receptor [Hylobates concolor] Length=275 (crested gibbon)
52 - 326
results alignment section
beta-2 adrenergic receptor [Ateles fusciceps] Length=275 (spider monkey)
52 - 326
results alignment section
52 - 326
The match is happening in the 7tm_1 region of the sequence which appears to be highly conserved.
| Hints: | Use the protein accession code from the previous exercise and run a protein BLAST search (http://blast.ncbi.nlm.nih.gov). This time, instead of using the default database, use the swissprotein database and a structure database. Compare the available structure information to make the decision. |
| transmembrane segments: | 1. 35 - 58 | 3. 107 - 129 | 5. 197 - 220 | 7. 306 - 329 |
| 2. 72 - 95 | 4. 151 - 174 | 6. 275 - 298 |
| comparison - The results show that the human proteins being compared are identical to one another. However, the structure and the swissprotein TMD segments don't agree as to number and location. Perhaps more study needs to be done on this protein to get the correct TMD locations and a complete structure. |
#4 step by step instructions
| 1. | Go to http://www.ncbi.nlm.nih.gov, and from the blue navigation bar click on BLAST. |

| 2. | From the main BLAST page, click on protein blast in the Basic BLAST section. |
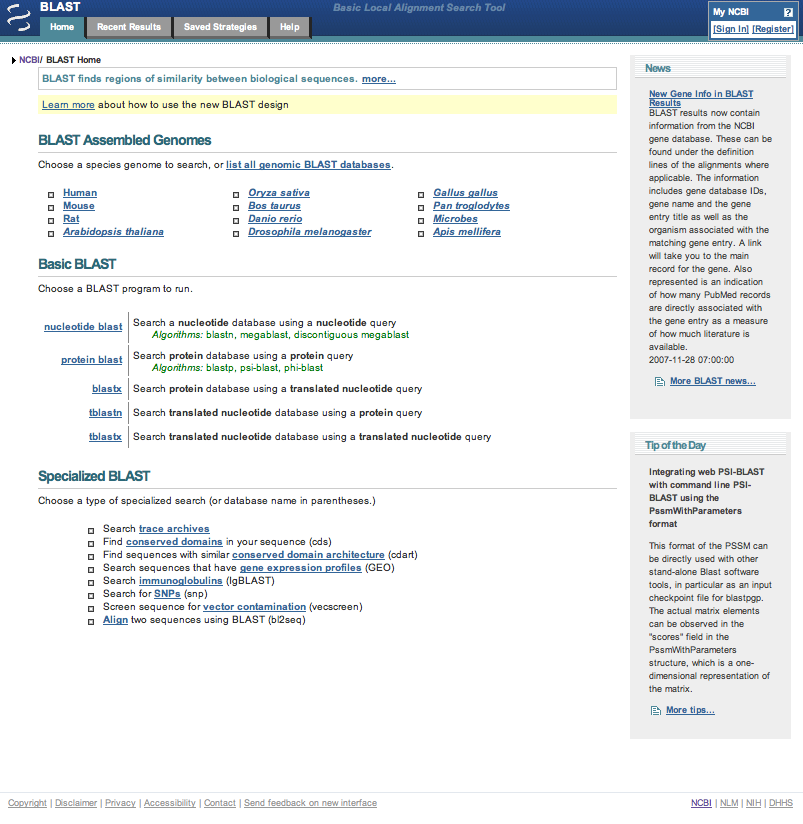
| In the blastp suite page, click on the ? icon at the end of the Database line in the Choose Search Set section to find out information about the databases that can be used in this protein BLAST search. Clicking on the more... link provides additional information. Once a suitable structure database name has been located, close the more... page and re-click on the "? icon" to close the information block. |
| From the list given the structural database to use is pdb (Protein Data Bank proteins). The swissprotein database (Swissprot protein sequences) was also listed. | |
| Of the protein databases, swissprotein is considered to have the best annotation. One of the features they report is transmembrane segment locations when available or predicted. | |
| 3. | Change the Choose Search Set Database option from nr to swissprotein using the pull down menu, |
| enter the previously found RefSeq protein accession code into the Enter Query Sequence box. |

| To speed things up and reduce the size of the output file, restrict the organism searched to humans by starting to enter homo sapiens in the Organism line. Select the proper line when it appears. |

| Start the run by clicking the BLAST button. |

| 4. | At the top of the actual results page, click on the "Reformat these Results" link. |

| This leads off to a form which allows the changing of the produced results. |
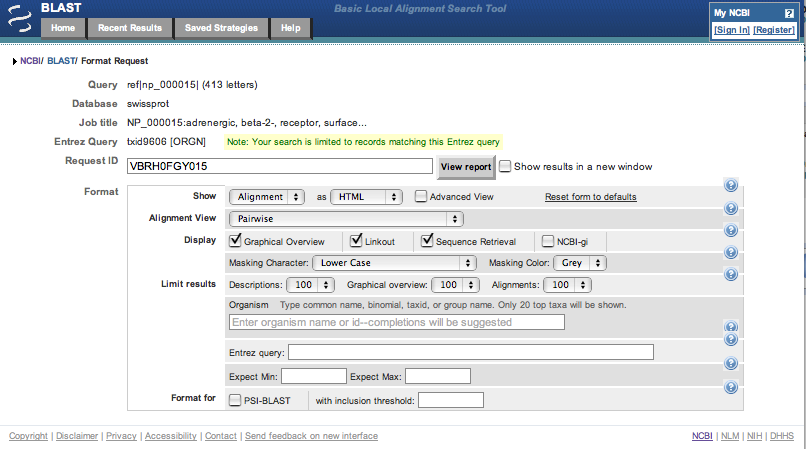
| The number of descriptions, lines in the image and alignments can be restricted using the Descriptions:, Graphical overview:
and Alignments: pull down menus. Restrict these three options to 10 each and then click on the View report button near the top of
the page. Here is the top part of the results page. |
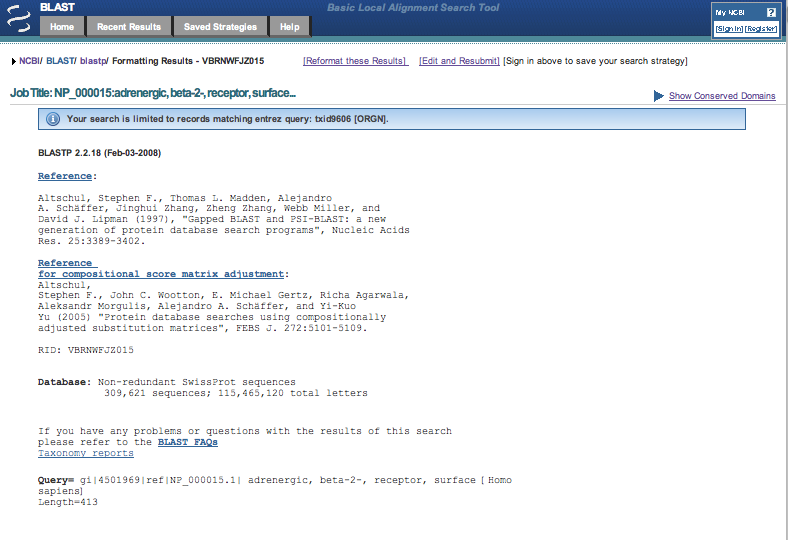

| 5. | Scroll down the results to the significant alignments section and click on the sequence link containing the term ADRB2_HUMAN. It should be the first one on the list and hist a 100% match to the submitted reference sequence. |
| link to ADRB2_human | |
| 6. | Scroll down the swissprotein data file to the FEATURES section. Then read through the listed features to find those regions called "Transmembrane region" and record them. |
| The first Transmembrane region from the data file |

|
|||||||||||
|
|||||||||||
| 7. | Return to the protein blast page, re-enter the RefSeq accession code if necessary, and change the database to be used to pdb, and return Organism to its default blank value. |

| Click BLAST button. | |
| 8. | Wait until results page appears. |
| 9. | The best hit comes from Human Beta2 Adrenoceptor and is a perfect match. |
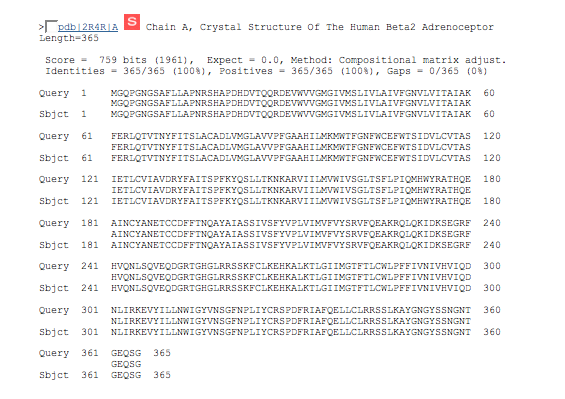
| The alignment does cover the entire area containing the transmembrane segments. The pdb code is in two parts, the first four alphanumeric characters refer to the structure name and the character after the | refers to the chain within the structure that has the match. |
| 10. | To find out more information about this structure, click on the red boxed S. This goes off to a "Related Structures" page. |
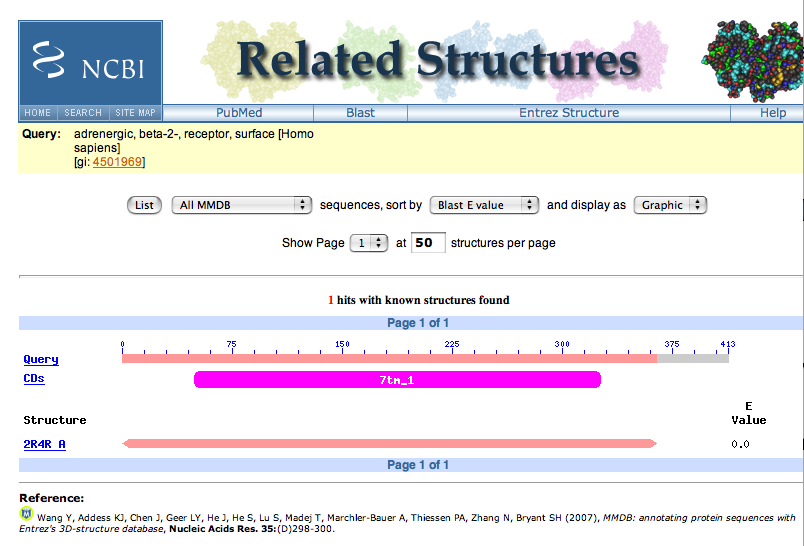
| The image shows that the 100% match is only for the first ~ 370 residues of the protein. Click on the name of the structure in the lower left hand side of the page. |
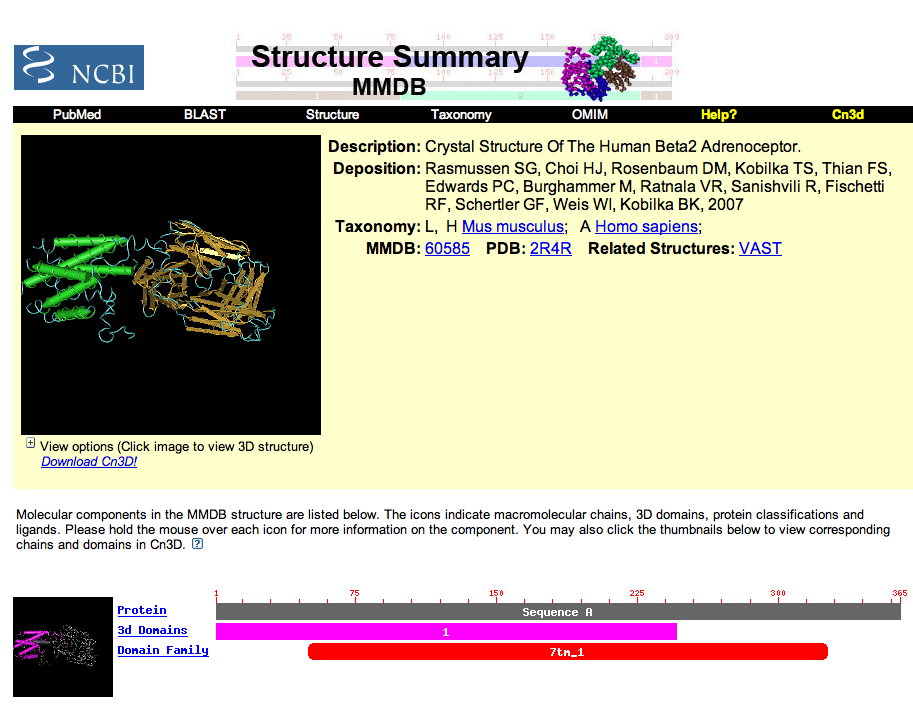
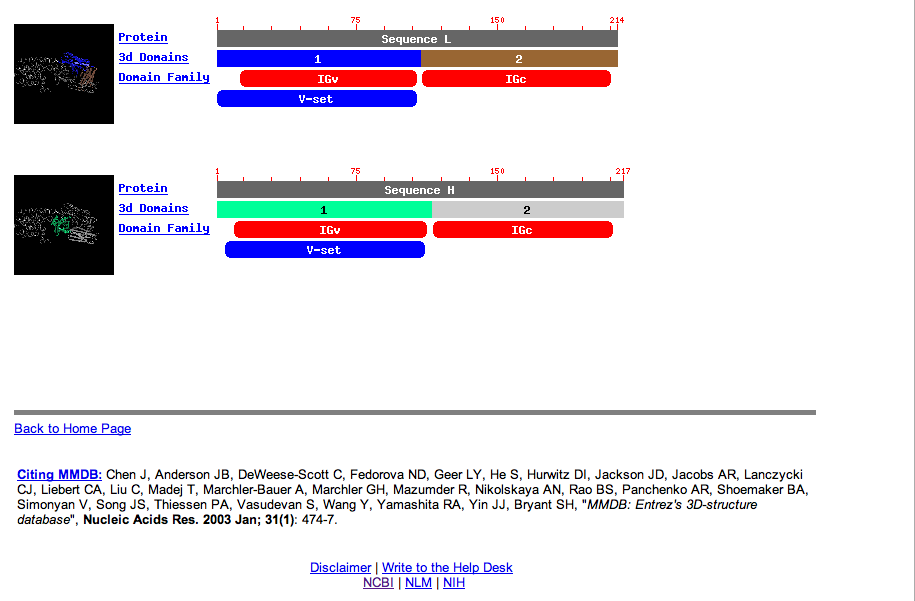
| Now at the Structure Summary site, more information is given about the actual structure. Sequence A (chain A) that matches the protein appears to have structural information for residues 1 to about 245, or the first five TMD sections according to swissprotein. There are two other parts of the structure which don't appear to be part of the ADBR2 protein. Click on the pdb link in the top section of the page to find out more. |
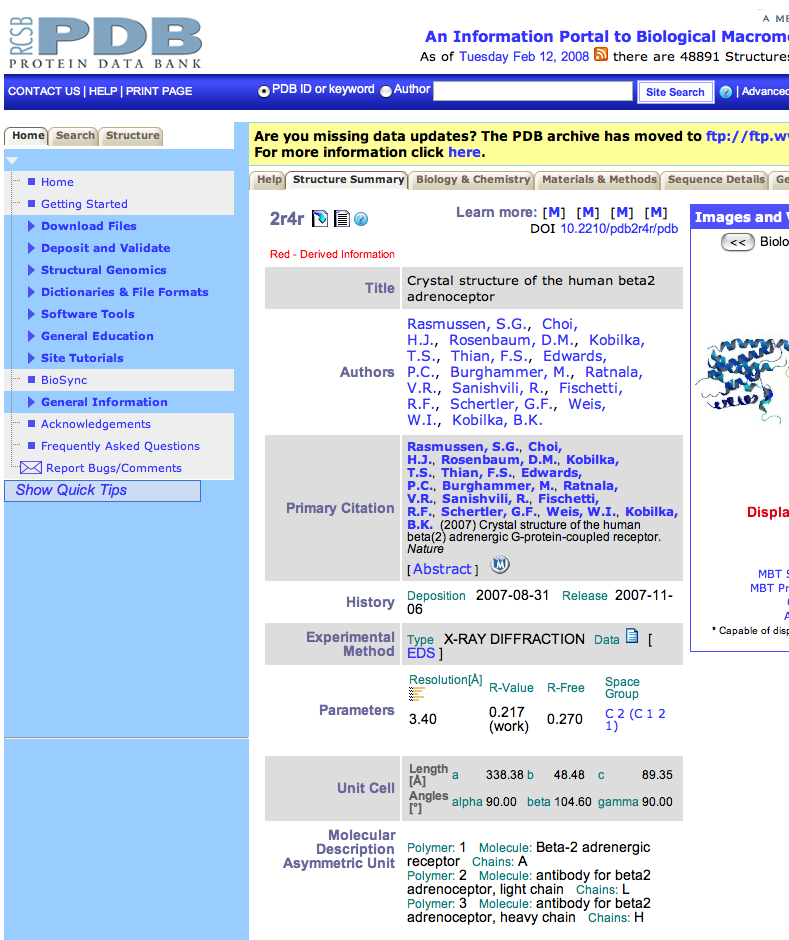
| Scrolling down the PDB page on this structure to the Molecular Description section
results in finding out that chains L and H are antibodies to human beta2 adrenoceptor protein.
Transmembrane proteins are very difficult to crystallize, the first step in doing x-ray
diffraction studies. It appears that attaching these antibodies to the protein made
crystallization possible. Return to the Structure Summary page and look closely at the green and gold image at the top of the page. The region with the green cylinders is the part of the structure from the ADRB2_HUMAN protein and the gold regions are the antibodies. Count the number of green cylinders shown. |
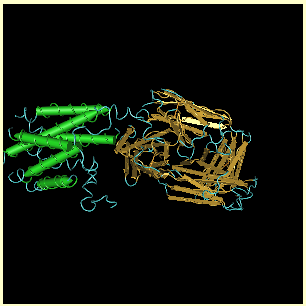
| There are 6 distinct cylinders. These represent helical structural elements in the protein. TMD sections for most proteins are expected to be helices. However, according to the swissprotein data, the part of the protein that was crystallized should only contain 5. Perhaps, additional study needs to be done on this protein to clarify the number of TMD sections the protein contains and where they are located. |
| Hints: | Determine the proteins associated with the disease by doing an Entrez protein search Choose the top hit and check its length. Use the sequence in a NCBI protein BLAST search to try to find possible model animals. Also use this sequence to find dog sequences. Compare any found dog sequence(s) with the human sequence you started with. |
| best dog match | NP_001006646 600 aa |
| complete dog sequence - Yes, but, there is a 15 residue gap starting at residue 72 that would have to be further investigated before proceeding. |
#5 step by step instructions
| 1. | Go to http://www.ncbi.nlm.nih.gov, change the All Databases option to Protein and enter the term pulmonary artery hypertension homo sapiens into the for box |

| and click Go. | |
| The results of the search. |
| 2. | Choose the top hit, in this case NP_002225. Clicking on the link will take you to the page for the
protein. |
| 3. | Check out the length of the protein. The length is the second item on the "LOCUS" line and is a number followed by aa. |
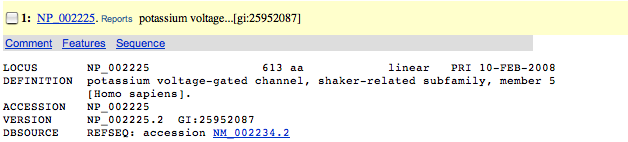
|
|
| (The sequence to be used in the search is 613 residues long.) | |
| Scan down the presented references to ensure that this protein has some relationship to pulmonary artery hypertension. |

| Reference 4 seems to indicate that SNPs in this protein were found in patients with idiopathic pulmonary arterial hypertension that impacted function. | |
| 4. | To obtain the sequence of NP_002225 for the BLAST search, change the Display option of the page from GenPept to FASTA using the pull down menu. |
| This automatically changes the format to FASTA. |
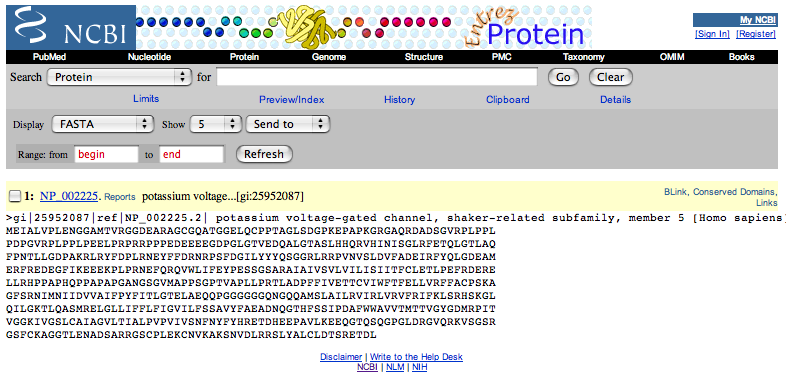
| Copy this data, starting with the ">" and continuing to the end of the sequence. |

| 5. | Click on the NCBI logo in the upper left-hand side of the screen. |
| 6. | From the main NCBI page click on BLAST in the blue navigation bar. |

| On the main BLAST page, click on protein blast in the Basic BLAST portion of the page. |

| 7. | Paste your sequence into the Enter Query Sequence box, |
| be sure that the Choose Search Set parameter are at their default values (database nr and organism blank) |
| and then click the BLAST button. | |
| 8. | Wait until the results page appears. |
| 9. | Check out the best hits with a description that appears to be correct, |
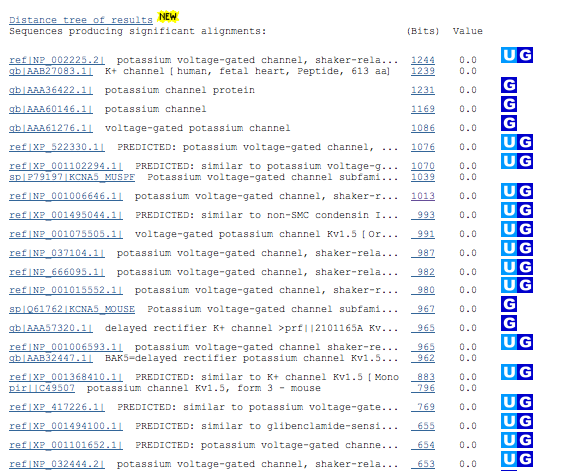
| Checking out the best hits to determine the quality of the matches and the species results in the following information. |
| XP_522330 | Pan troglodytes (chimpanzee) | 602 residues | Identities = 600/613 (97%) |
| XP_001102294 | Macaca mulatta (rhesus monkey) | 605 residues | Identities = 596/616 (96%) |
| KCNA5_MUSPF | Mustela putorius furo (domestic ferret) | 601 residues | Identities = 547/613 (89%) |
| NP_001006646 | Canis lupus familiaris (dog) | 600 residues | Identities = 538/615 (87%) |
| XP_001495044 | Equus caballus (horse) | 595 residues | Identities = 539/614 (87%) |
| NP_001075505 | Oryctolagus cuniculus (rabbit) | 598 residues | Identities = 533/616 (86%) |
| NP_037104 | Rattus norvegicus (Norway rat) | 602 residues | Identities = 529/615 (86%) |
| NP_666095 | Mus musculus (house mouse) | 602 residues | Identities = 530/615 (86%) |
| NP_001015552 | Bos taurus (cattle) | 598 residues | Identities = 526/614 (85%) |
| NP_001006593 | Sus scrofa (pig) | 600 residues | Identities = 524/616 (85%) |
| XP_001368410 | Monodelphis domestica (gray short-tailed opossum) | 609 residues | Identities = 472/621 (76%) |
| These results would indicate that rhesus monkey [Macaca mulatta] would be the best model. However, dog, rabbit, mouse and rat would all be good animal models in which to study this gene and its function. | |
| 10. | Return to the blastp suite submission page and change the Organism option in the Choose Search Set section from blank to Canis familiaris by starting to enter this term into the field. Highlight the term when it appears on the list. |

| Click the BLAST button to start the run. |

| Confirm that the hits are from dog and determine how close the length is to that of the starting human sequence. The first one looks the most likely. |
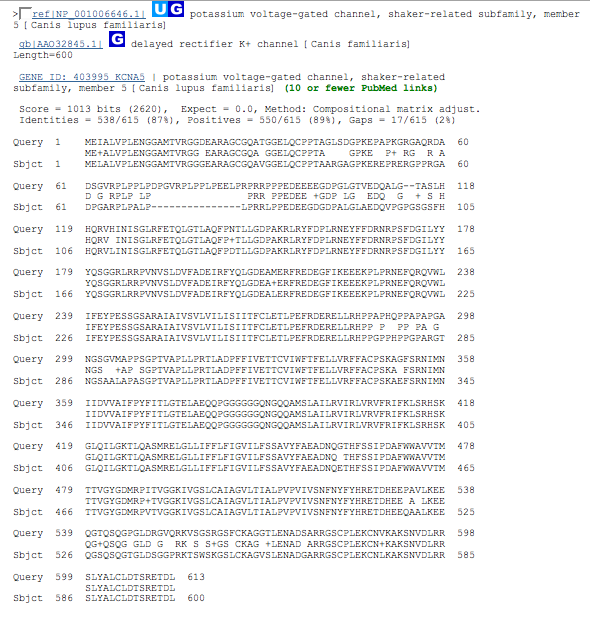
| Looking at these results there is only one real area of concern. The 15 residue gap starting at position 72 would need to be looked at to find out if there are any known functions or features associated with this region in the human sequence. If so, then the dog protein wouldn't be a good study model. |
| #6 | Are there knockout mice available to study the AGPAT6 gene? How would you order one of these cell lines? |
| Hints: | Find the mRNA FASTA formatted sequence for the AGPAT6 mouse gene by doing an Entrez Gene search at NCBI (http://www.ncbi.nlm.nih.gov). Then a BLAST search at the International Gene Trap Consortium (IGTC) site (http://www.genetrap.org) to see if such knockouts exist. |
#6 step by step instructions
| 1. | Go to http://www.ncbi.nlm.nih.gov, change the Search option to Gene, enter AGPAT6 in the for box |

| and click Go. |

| 2. | Click on the mouse link on the search results page to go mouse Entrez Gene page. Scroll down this page to the NCBI Reference Sequences (RefSeq) section. |
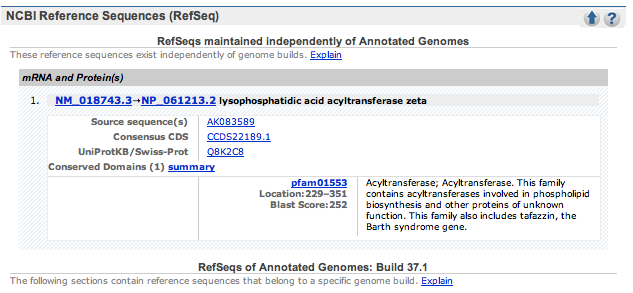
Click on the mRNA sequence link.
|
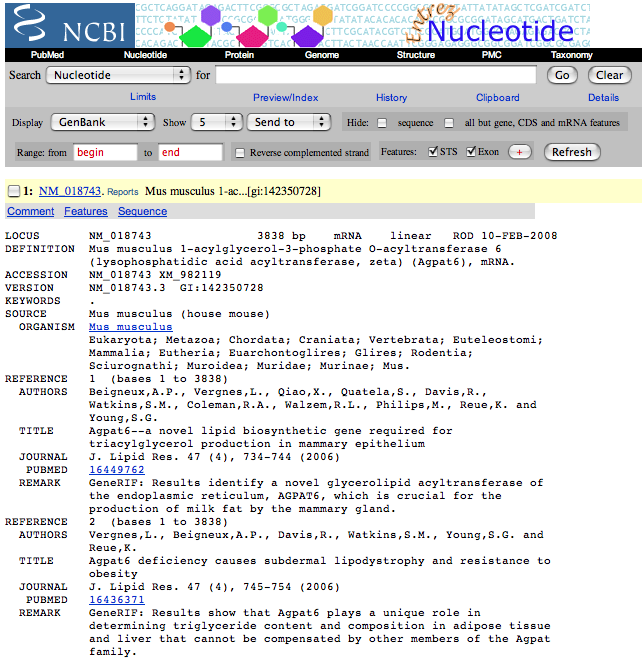
| 3. | Convert the sequence in the default format to a FASTA formatted file by changing the Display option to FASTA. Copy this sequence. |

| 4. | Go to the IGTC web site (http://www.genetrap.org). |
| Left side of the IGTC home page. News items on the right side change with time | |
| Click on DATA ACCESS in the blue navigation bar to see the options available. Select the Blast Search option. |
| 5. | Paste your mRNA sequence into the Enter sequence below box |
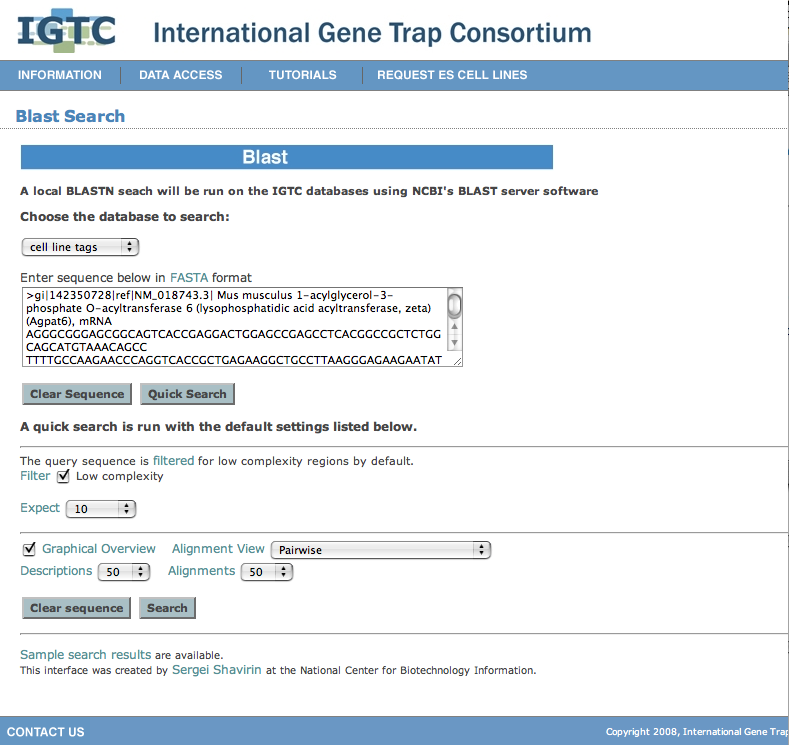
| and click Quick Search. |
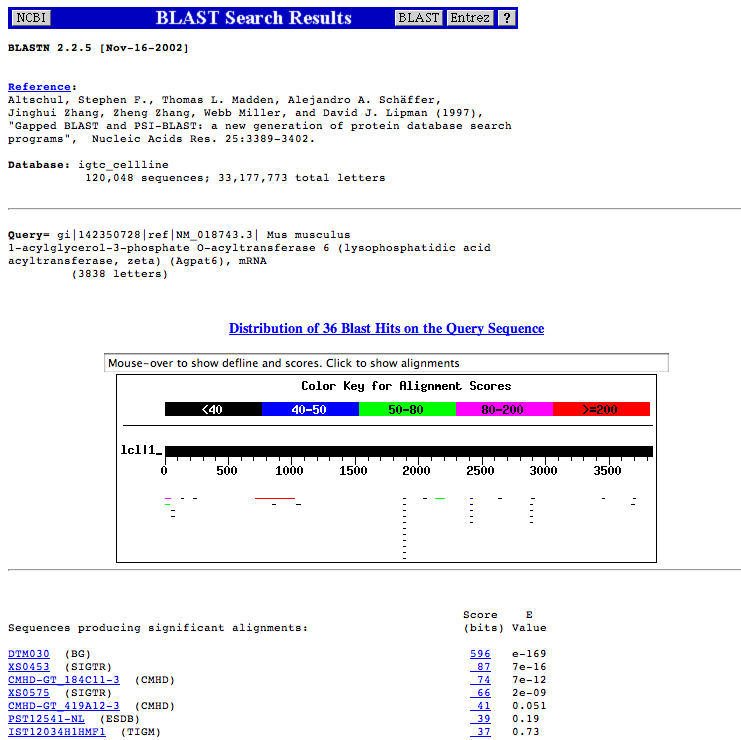
| Using an mRNA sequence for a Blast search at this site allows the detection of standard loss-of-function allele cell lines that have a match with the mRNA sequence. To find intronic ones would require the use of the sequence for the genomic region occupied by the mRNA sequence. | |
| 6. | Scroll through the results and look at the actual alignments. |


| The IGTC web site uses strict guidelines for associating a cell line with a gene. A match needs to be at least 50%
of the cell line length and have an identity of at least 90%. Using this criteria the following three cell lines are associated with the AGPAT6 gene:
|
|
| 7. | Click on one of these cell line links to go off to a cell line annotation page. Here data is presented on the cell line, the gene it is associated with, and an image is given displaying the location of the cell line with respect to the gene's mRNA sequence. |
| To see all the data that is available on this page, click the Show All arrow in the Additional Information. To hide this information again, click on Hide All. |
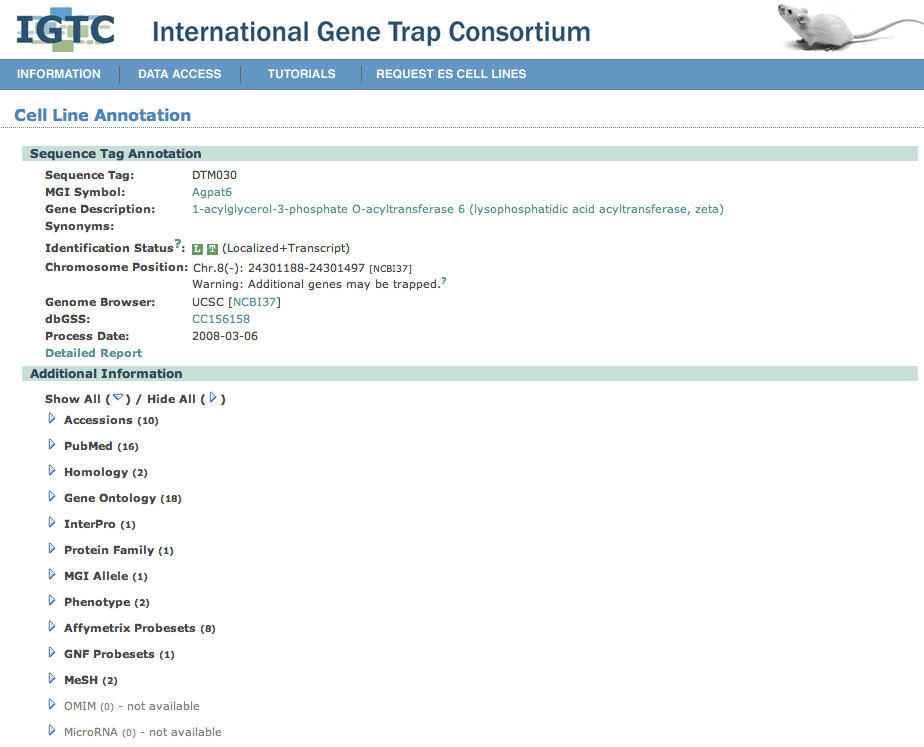
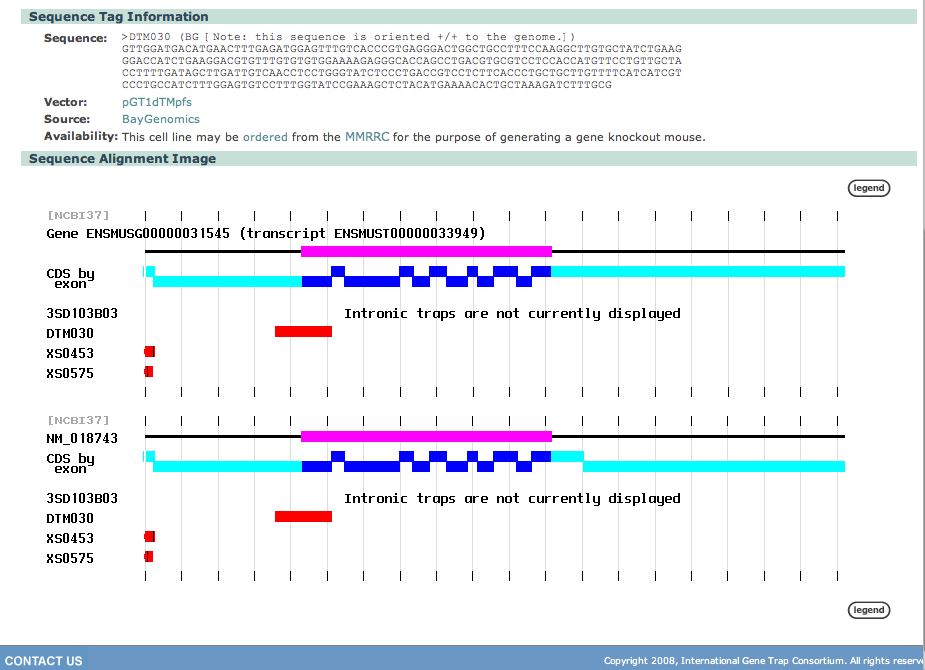
| In the Sequence Tag Information section of the page, |

information is provided on the source of the cell line and how to order it via a provided link.
order from MMRRC
XS0453 and XS0575 are from the Sanger International Gene Trap Resource order from the MMRRC
|
|
| Clicking on the Gene Description link goes off to a Gene Annotation page. |
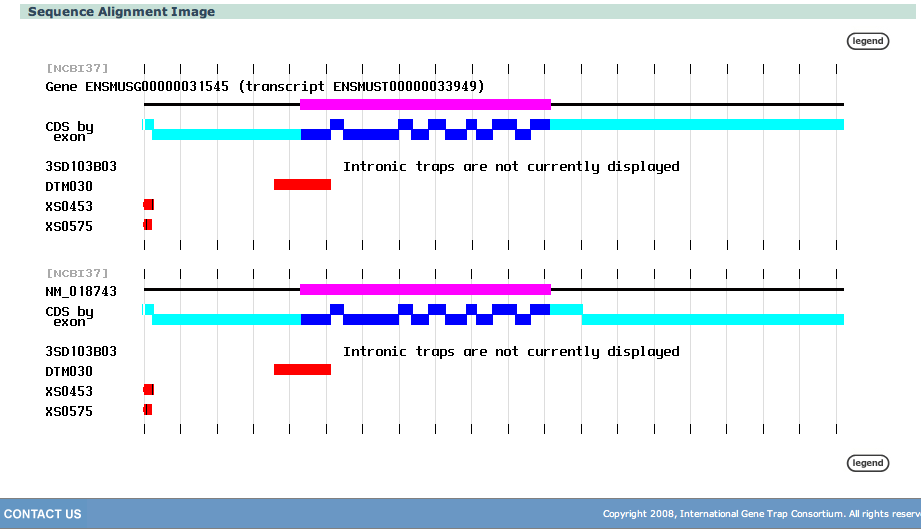
| The image shows that cell lines XS0453 and XS0575 occur in approximately the same place, while DTM030 is further down stream and it is impossible to figure out where the intronic trap may be. | |
| There is another way to determine this information at the site and that is to do a gene search with the gene name. For more details on doing this check out the Search tutorial in the TUTORIALS pull down menu. This technique, however, doesn't allow the user to assess the quality of the gene matches. |
last updated 5/30/2008